Regulation of Notch output dynamics via specific E(spl)-HLH factors during bristle patterning in Drosophila
- PMID: 31375669
- PMCID: PMC6677740
- DOI: 10.1038/s41467-019-11477-2
Regulation of Notch output dynamics via specific E(spl)-HLH factors during bristle patterning in Drosophila
Abstract
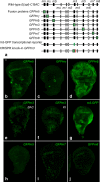
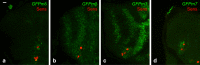
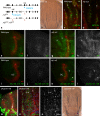
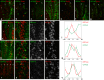
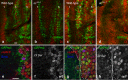
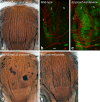
The stereotyped arrangement of sensory bristles on the adult fly thorax arises from a self-organized process, in which inhibitory Notch signaling both delimits proneural stripes and singles out sensory organ precursor cells (SOPs). A dynamic balance between proneural factors and Enhancer of split-HLH (E(spl)-HLH) Notch targets underlies patterning, but how this is regulated is unclear. Here, were identify two classes of E(spl)-HLH factors, whose expression both precedes and delimits proneural activity, and is dependent on proneural activity and required for proper SOP spacing within the stripes, respectively. These two classes are partially redundant, since a member of the second class, that is normally cross-repressed by members of the first class, can functionally compensate for their absence. The regulation of specific E(spl)-HLH genes by proneural factors amplifies the response to Notch as SOPs are being selected, contributing to patterning dynamics in the notum, and likely operates in other developmental contexts.
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
A re-examination of the selection of the sensory organ precursor of the bristle sensilla of Drosophila melanogaster.PLoS Genet. 2015 Jan 8;11(1):e1004911. doi: 10.1371/journal.pgen.1004911. eCollection 2015 Jan. PLoS Genet. 2015. PMID: 25569355 Free PMC article.
-
Self-organized Notch dynamics generate stereotyped sensory organ patterns in Drosophila.Science. 2017 May 5;356(6337):eaai7407. doi: 10.1126/science.aai7407. Epub 2017 Apr 6. Science. 2017. PMID: 28386027
-
Hairless promotes stable commitment to the sensory organ precursor cell fate by negatively regulating the activity of the Notch signaling pathway.Dev Biol. 1995 Dec;172(2):479-94. doi: 10.1006/dbio.1995.8033. Dev Biol. 1995. PMID: 8612965
-
Morphogenesis of Drosophila melanogaster macrochaetes: cell fate determination for bristle organ.J Stem Cells. 2012;7(1):19-41. J Stem Cells. 2012. PMID: 23550342 Review.
-
Evolution of cis-regulation of the proneural genes.Int J Dev Biol. 2003;47(7-8):643-51. Int J Dev Biol. 2003. PMID: 14756340 Review.
Cited by
-
Evolution and genomic basis of the plant-penetrating ovipositor: a key morphological trait in herbivorous Drosophilidae.Proc Biol Sci. 2022 Nov 9;289(1986):20221938. doi: 10.1098/rspb.2022.1938. Epub 2022 Nov 9. Proc Biol Sci. 2022. PMID: 36350206 Free PMC article.
-
Dissecting, Fixing, and Visualizing the Drosophila Pupal Notum.J Vis Exp. 2022 Apr 6;(182):10.3791/63682. doi: 10.3791/63682. J Vis Exp. 2022. PMID: 35467663 Free PMC article.
-
Sequential and independent probabilistic events regulate differential axon targeting during development in Drosophila melanogaster.Nat Neurosci. 2025 May;28(5):998-1011. doi: 10.1038/s41593-025-01937-y. Epub 2025 May 7. Nat Neurosci. 2025. PMID: 40335773
-
High-Temporal-Resolution smFISH Method for Gene Expression Studies in Caenorhabditis elegans Embryos.Anal Chem. 2021 Jan 26;93(3):1369-1376. doi: 10.1021/acs.analchem.0c02966. Epub 2020 Dec 23. Anal Chem. 2021. PMID: 33355449 Free PMC article.
-
A combination of Notch signaling, preferential adhesion and endocytosis induces a slow mode of cell intercalation in the Drosophila retina.Development. 2021 May 15;148(10):dev197301. doi: 10.1242/dev.197301. Epub 2021 May 17. Development. 2021. PMID: 33999996 Free PMC article.
References
-
- Held LI. Imaginal Discs: the Genetic and Cellular Logic of Pattern Formation. New York: Cambridge University Press, Cambridge; 2002.
-
- Hartenstein V, Posakony JW. Development of adult sensilla on the wing and notum of Drosophila melanogaster. Development. 1989;107:389–405. - PubMed
-
- Gho M, Bellaiche Y, Schweisguth F. Revisiting the Drosophila microchaete lineage: a novel intrinsically asymmetric cell division generates a glial cell. Development. 1999;126:3573–3584. - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

