Keratin 17 identifies the most lethal molecular subtype of pancreatic cancer
- PMID: 31375762
- PMCID: PMC6677817
- DOI: 10.1038/s41598-019-47519-4
Keratin 17 identifies the most lethal molecular subtype of pancreatic cancer
Abstract
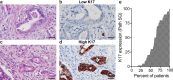
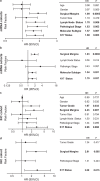
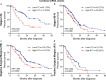
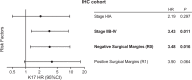
Although the overall five-year survival of patients with pancreatic ductal adenocarcinoma (PDAC) is dismal, there are survival differences between cases with clinically and pathologically indistinguishable characteristics, suggesting that there are uncharacterized properties that drive tumor progression. Recent mRNA sequencing studies reported gene-expression signatures that define PDAC molecular subtypes that correlate with differences in survival. We previously identified Keratin 17 (K17) as a negative prognostic biomarker in other cancer types. Here, we set out to determine if K17 is as accurate as molecular subtyping of PDAC to identify patients with the shortest survival. K17 mRNA was analyzed in two independent PDAC cohorts for discovery (n = 124) and validation (n = 145). Immunohistochemical localization and scoring of K17 immunohistochemistry (IHC) was performed in a third independent cohort (n = 74). Kaplan-Meier and Cox proportional-hazard regression models were analyzed to determine cancer specific survival differences in low vs. high mRNA K17 expressing cases. We established that K17 expression in PDACs defines the most aggressive form of the disease. By using Cox proportional hazard ratio, we found that increased expression of K17 at the IHC level is also associated with decreased survival of PDAC patients. Additionally, within PDACs of advanced stage and negative surgical margins, K17 at both mRNA and IHC level is sufficient to identify the subgroup with the shortest survival. These results identify K17 as a novel negative prognostic biomarker that could inform patient management decisions.
Conflict of interest statement
L.F. Escobar-Hoyos and K.R. Shroyer are consultants for KDx Diagnostics Inc. and OncoGenesis Inc.
Figures


References
-
- Kamarajah SK, Burns WR, Frankel TL, Cho CS, Nathan H. Validation of the American Joint Commission on Cancer (AJCC) 8th Edition Staging System for Patients with Pancreatic Adenocarcinoma: A Surveillance, Epidemiology and End Results (SEER) Analysis. Ann Surg Oncol. 2017;24:2023–2030. doi: 10.1245/s10434-017-5810-x. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

