Compendium of 4,941 rumen metagenome-assembled genomes for rumen microbiome biology and enzyme discovery
- PMID: 31375809
- PMCID: PMC6785717
- DOI: 10.1038/s41587-019-0202-3
Compendium of 4,941 rumen metagenome-assembled genomes for rumen microbiome biology and enzyme discovery
Abstract
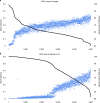
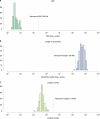
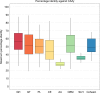
Ruminants provide essential nutrition for billions of people worldwide. The rumen is a specialized stomach that is adapted to the breakdown of plant-derived complex polysaccharides. The genomes of the rumen microbiota encode thousands of enzymes adapted to digestion of the plant matter that dominates the ruminant diet. We assembled 4,941 rumen microbial metagenome-assembled genomes (MAGs) using approximately 6.5 terabases of short- and long-read sequence data from 283 ruminant cattle. We present a genome-resolved metagenomics workflow that enabled assembly of bacterial and archaeal genomes that were at least 80% complete. Of note, we obtained three single-contig, whole-chromosome assemblies of rumen bacteria, two of which represent previously unknown rumen species, assembled from long-read data. Using our rumen genome collection we predicted and annotated a large set of rumen proteins. Our set of rumen MAGs increases the rate of mapping of rumen metagenomic sequencing reads from 15% to 50-70%. These genomic and protein resources will enable a better understanding of the structure and functions of the rumen microbiota.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Gerber, P. J et al. Tackling Climate Change Through Livestock: a Global Assessment of Emissions and Mitigation Opportunities. (Food and Agriculture Organization of the United Nations (FAO), 2013).
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

