Visualization of protein sequence space with force-directed graphs, and their application to the choice of target-template pairs for homology modelling
- PMID: 31377535
- PMCID: PMC7110651
- DOI: 10.1016/j.jmgm.2019.07.014
Visualization of protein sequence space with force-directed graphs, and their application to the choice of target-template pairs for homology modelling
Abstract
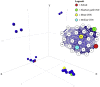
The protein sequence-structure gap results from the contrast between rapid, low-cost deep sequencing, and slow, expensive experimental structure determination techniques. Comparative homology modelling may have the potential to close this gap by predicting protein structure in target sequences using existing experimentally solved structures as templates. This paper presents the first use of force-directed graphs for the visualization of sequence space in two dimensions, and applies them to the choice of suitable RNA-dependent RNA polymerase (RdRP) target-template pairs within human-infective RNA virus genera. Measures of centrality in protein sequence space for each genus were also derived and used to identify centroid nearest-neighbour sequences (CNNs) potentially useful for production of homology models most representative of their genera. Homology modelling was then carried out for target-template pairs in different species, different genera and different families, and model quality assessed using several metrics. Reconstructed ancestral RdRP sequences for individual genera were also used as templates for the production of ancestral RdRP homology models. High quality ancestral RdRP models were consistently produced, as were good quality models for target-template pairs in the same genus. Homology modelling between genera in the same family produced mixed results and inter-family modelling was unreliable. We present a protocol for the production of optimal RdRP homology models for use in further experiments, e.g. docking to discover novel anti-viral compounds. (219 words).
Keywords: Force-directed graphs; Fruchterman-Reingold algorithm; Homology modelling; Multidimensional scaling; Protein structure; RNA-Dependent RNA polymerase; Reverse transcriptase; Sequence space; Sequence-structure gap.
Copyright © 2019 Elsevier Inc. All rights reserved.
Figures

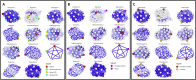


 ), allowed (
), allowed ( ), and outliers (
), and outliers ( cross,
cross,  text). The Z-score graphics show model quality on a sliding scale: low-quality (
text). The Z-score graphics show model quality on a sliding scale: low-quality ( ), high-quality (
), high-quality ( ). QMEAN4 shows the overall Z-score, “All Atom” shows the average Z-score for all of the atoms in the model, “CBeta” the Z-score for all Cβ carbons, “Solvation” is a measure of how accessible the residues are to solvents, and “Torsion” is a measure of torsion angle for each residue compared to adjacent residues.
). QMEAN4 shows the overall Z-score, “All Atom” shows the average Z-score for all of the atoms in the model, “CBeta” the Z-score for all Cβ carbons, “Solvation” is a measure of how accessible the residues are to solvents, and “Torsion” is a measure of torsion angle for each residue compared to adjacent residues.
Similar articles
-
Space constrained homology modelling: the paradigm of the RNA-dependent RNA polymerase of dengue (type II) virus.Comput Math Methods Med. 2013;2013:108910. doi: 10.1155/2013/108910. Epub 2013 Aug 6. Comput Math Methods Med. 2013. PMID: 23986788 Free PMC article.
-
Functional insights from molecular modeling, docking, and dynamics study of a cypoviral RNA dependent RNA polymerase.J Mol Graph Model. 2015 Sep;61:160-74. doi: 10.1016/j.jmgm.2015.07.002. Epub 2015 Jul 26. J Mol Graph Model. 2015. PMID: 26264734
-
An evaluation of automated homology modelling methods at low target template sequence similarity.Bioinformatics. 2007 Aug 1;23(15):1901-8. doi: 10.1093/bioinformatics/btm262. Epub 2007 May 17. Bioinformatics. 2007. PMID: 17510171
-
A guide to template based structure prediction.Curr Protein Pept Sci. 2009 Jun;10(3):270-85. doi: 10.2174/138920309788452182. Curr Protein Pept Sci. 2009. PMID: 19519455 Review.
-
Bridging the gap between structural bioinformatics and receptor research: the membrane-embedded, ligand-gated, P2X glycoprotein receptor.Curr Top Med Chem. 2004;4(16):1657-705. doi: 10.2174/1568026043387197. Curr Top Med Chem. 2004. PMID: 15579102 Review.
References
-
- Kendrew J.C., Bodo G., Dintzis H.M., Parrish R.G., Wyckoff H., Phillips D.C. A three-dimensional model of the myoglobin molecule obtained by x-ray analysis. Nature. 1958;181:662–666. - PubMed
-
- Holmes E.C. Oxford University Press; Oxford, UK: 2009. The Evolution and Emergence of RNA Viruses.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

