MISTERMINATE Mechanistically Links Mitochondrial Dysfunction with Proteostasis Failure
- PMID: 31378462
- PMCID: PMC7362879
- DOI: 10.1016/j.molcel.2019.06.031
MISTERMINATE Mechanistically Links Mitochondrial Dysfunction with Proteostasis Failure
Abstract
Mitochondrial dysfunction and proteostasis failure frequently coexist as hallmarks of neurodegenerative disease. How these pathologies are related is not well understood. Here, we describe a phenomenon termed MISTERMINATE (mitochondrial-stress-induced translational termination impairment and protein carboxyl terminal extension), which mechanistically links mitochondrial dysfunction with proteostasis failure. We show that mitochondrial dysfunction impairs translational termination of nuclear-encoded mitochondrial mRNAs, including complex-I 30kD subunit (C-I30) mRNA, occurring on the mitochondrial surface in Drosophila and mammalian cells. Ribosomes stalled at the normal stop codon continue to add to the C terminus of C-I30 certain amino acids non-coded by mRNA template. C-terminally extended C-I30 is toxic when assembled into C-I and forms aggregates in the cytosol. Enhancing co-translational quality control prevents C-I30 C-terminal extension and rescues mitochondrial and neuromuscular degeneration in a Parkinson's disease model. These findings emphasize the importance of efficient translation termination and reveal unexpected link between mitochondrial health and proteome homeostasis mediated by MISTERMINATE.
Keywords: CAT-tailing; MISTERMINATE; PINK1/Parkin; Parkinson’s disease; RQC; mitochondrial stress; neurodegeneration; proteostasis; ribosome stalling; translation termination.
Copyright © 2019 Elsevier Inc. All rights reserved.
Conflict of interest statement
DECLARATION OF INTERESTS
The authors declare no competing interests.
Figures




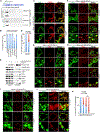
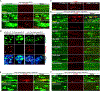
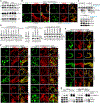
References
-
- Bi X, Jones T, Abbasi F, Lee H, Stultz B, Hursh DA, and Mortin MA (2005). Drosophila caliban, a nuclear export mediator, can function as a tumor suppressor in human lung cancer cells. Oncogene 24, 8229–8239. - PubMed
-
- Bossy-Wetzel E, Schwarzenbacher R, and Lipton SA (2004). Molecular pathways to neurodegeneration. Nat. Med 10 (Suppl), S2–S9. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

