Actin Dynamics Drive Microvillar Motility and Clustering during Brush Border Assembly
- PMID: 31378589
- PMCID: PMC6736749
- DOI: 10.1016/j.devcel.2019.07.008
Actin Dynamics Drive Microvillar Motility and Clustering during Brush Border Assembly
Abstract
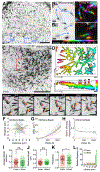
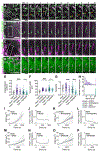
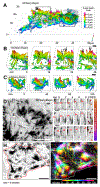
Transporting epithelial cells generate arrays of microvilli, known as a brush border, to enhance functional capacity. To understand brush border formation, we used live cell imaging to visualize apical remodeling early in this process. Strikingly, we found that individual microvilli exhibit persistent active motility, translocating across the cell surface at ∼0.2 μm/min. Perturbation with inhibitors and photokinetic experiments revealed that microvillar motility is driven by actin assembly at the barbed ends of core bundles, which in turn is linked to robust treadmilling of these structures. Actin regulatory factors IRTKS and EPS8 localize to the barbed ends of motile microvilli, where they control the kinetics and nature of movement. As the apical surface of differentiating epithelial cells is crowded with nascent microvilli, persistent motility promotes collisions between protrusions and ultimately clustering and consolidation into higher-order arrays. Thus, microvillar motility represents a previously unrecognized driving force for apical surface remodeling and maturation during epithelial differentiation.
Keywords: adhesion; bundle; epithelium; protocadherin; treadmilling.
Copyright © 2019 Elsevier Inc. All rights reserved.
Conflict of interest statement
DECLARATION OF INTERESTS
The authors declare no competing interests.
Figures



Comment in
-
Moving Encounters: Actin Treadmilling in the Brush Border.Dev Cell. 2019 Sep 9;50(5):529-530. doi: 10.1016/j.devcel.2019.08.011. Dev Cell. 2019. PMID: 31505173
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

