Cancer Cells Upregulate NRF2 Signaling to Adapt to Autophagy Inhibition
- PMID: 31378590
- PMCID: PMC7233142
- DOI: 10.1016/j.devcel.2019.07.010
Cancer Cells Upregulate NRF2 Signaling to Adapt to Autophagy Inhibition
Abstract
While autophagy is thought to be an essential process in some cancer cells, it is unknown if or how such cancer cells can circumvent autophagy inhibition. To address this, we developed a CRISPR/Cas9 assay with dynamic live-cell imaging to measure acute effects of knockout (KO) of autophagy genes compared to known essential and non-essential genes. In some cancer cells, autophagy is as essential for cancer cell growth as mRNA transcription or translation or DNA replication. However, even these highly autophagy-dependent cancer cells evolve to circumvent loss of autophagy by upregulating NRF2, which is necessary and sufficient for autophagy-dependent cells to circumvent ATG7 KO and maintain protein homeostasis. Importantly, however, this adaptation increases susceptibly to proteasome inhibitors. These studies identify a common mechanism of acquired resistance to autophagy inhibition and show that selection to avoid tumor cell dependency on autophagy creates new, potentially actionable cancer cell susceptibilities.
Keywords: ATG7; CRISPR/Cas9; NRF2; autophagy; cancer; chloroquine resistance; oxidative stress; proteasomal degradation.
Copyright © 2019. Published by Elsevier Inc.
Conflict of interest statement
DECLARATION OF INTERESTS
The authors declare no competing interests.
Figures


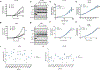

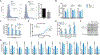
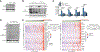

References
-
- Alam J, Killeen E, Gong P, Naquin R, Hu B, Stewart D, Ingelfinger JR, and Nath KA (2003). Heme activates the heme oxygenase-1 gene in renal epithelial cells by stabilizing Nrf2. Am. J. Physiol. Renal Physiol 284, F743–F752. - PubMed
-
- Arlt A, Bauer I, Schafmayer C, Tepel J, Mu€erkö ster SS, Brosch M, Rö der C, Kalthoff H, Hampe J, Moyer MP, et al. (2009). Increased proteasome subunit protein expression and proteasome activity in colon cancer relate to an enhanced activation of nuclear factor E2-related factor 2 (Nrf2). Oncogene 28, 3983–3996. - PubMed
-
- Blomen VA, Má jek P, Jae LT, Bigenzahn JW, Nieuwenhuis J, Staring J, Sacco R, Van Diemen FR, Olk N, Stukalov A, et al. (2015). Gene essentiality and synthetic lethality in haploid human cells. Science 350, 1092–1096. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

