Mapping Interactions of Microbial Metabolites with Human G-Protein-Coupled Receptors
- PMID: 31378678
- PMCID: PMC6706627
- DOI: 10.1016/j.chom.2019.07.002
Mapping Interactions of Microbial Metabolites with Human G-Protein-Coupled Receptors
Abstract
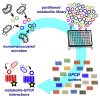
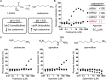
Despite evidence linking the human microbiome to health and disease, how the microbiota affects human physiology remains largely unknown. Microbiota-encoded metabolites are expected to play an integral role in human health. Therefore, assigning function to these metabolites is critical to understanding these complex interactions and developing microbiota-inspired therapies. Here, we use large-scale functional screening of molecules produced by individual members of a simplified human microbiota to identify bacterial metabolites that agonize G-protein-coupled receptors (GPCRs). Multiple metabolites, including phenylpropanoic acid, cadaverine, 9-10-methylenehexadecanoic acid, and 12-methyltetradecanoic acid, were found to interact with GPCRs associated with diverse functions within the nervous and immune systems, among others. Collectively, these metabolite-receptor pairs indicate that diverse aspects of human health are potentially modulated by structurally simple metabolites arising from primary bacterial metabolism.
Keywords: G protein-coupled receptors; human microbiome; primary metabolites.
Copyright © 2019 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
S.F.B. is the founder of LODO Therapeutics. Patents related to this work are being filed.
Figures





Comment in
-
The Gut Feeling: GPCRs Enlighten the Way.Cell Host Microbe. 2019 Aug 14;26(2):160-162. doi: 10.1016/j.chom.2019.07.018. Cell Host Microbe. 2019. PMID: 31415748 Free PMC article.
References
-
- Ahmed K., Tunaru S., Langhans C.D., Hanson J., Michalski C.W., Kölker S., Jones P.M., Okun J.G., Offermanns S. Deorphanization of GPR109B as a receptor for the β-oxidation intermediate 3-OH-octanoic acid and its role in the regulation of lipolysis. J. Biol. Chem. 2009;284:21928–21933. - PMC - PubMed
-
- Bhattarai Y., Williams B.B., Battaglioli E.J., Whitaker W.R., Till L., Grover M., Linden D.R., Akiba Y., Kandimalla K.K., Zachos N.C. Gut microbiota-produced tryptamine activates an epithelial G-protein-coupled receptor to increase colonic secretion. Cell Host Microbe. 2018;23:775–785. - PMC - PubMed
-
- Brighton P.J., Szekeres P.G., Willars G.B. Neuromedin U and its receptors: structure, function, and physiological roles. Pharmacol. Rev. 2004;56:231–248. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

