CtIP is essential for telomere replication
- PMID: 31378812
- PMCID: PMC6755089
- DOI: 10.1093/nar/gkz652
CtIP is essential for telomere replication
Abstract
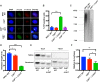
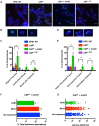
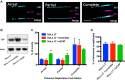
The maintenance of telomere length is critical to longevity and survival. Specifically, the failure to properly replicate, resect, and/or form appropriate telomeric structures drives telomere shortening and, in turn, genomic instability. The endonuclease CtIP is a DNA repair protein that is well-known to promote genome stability through the resection of endogenous DNA double-stranded breaks. Here, we describe a novel role for CtIP. We show that in the absence of CtIP, human telomeres shorten rapidly to non-viable lengths. This telomere dysfunction results in an accumulation of fusions, breaks, and frank telomere loss. Additionally, CtIP suppresses the generation of circular, extrachromosomal telomeric DNA. These latter structures appear to arise from arrested DNA replication forks that accumulate in the absence of CtIP. Hence, CtIP is required for faithful replication through telomeres via its roles at stalled replication tracts. Our findings demonstrate a new role for CtIP as a protector of human telomere integrity.
© The Author(s) 2019. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures




Similar articles
-
MAD2L2 controls DNA repair at telomeres and DNA breaks by inhibiting 5' end resection.Nature. 2015 May 28;521(7553):537-540. doi: 10.1038/nature14216. Epub 2015 Mar 23. Nature. 2015. PMID: 25799990 Free PMC article.
-
BRCA1 and CtIP promote alternative non-homologous end-joining at uncapped telomeres.EMBO J. 2015 Feb 3;34(3):410-24. doi: 10.15252/embj.201488947. Epub 2015 Jan 12. EMBO J. 2015. PMID: 25582120 Free PMC article.
-
CtIP-Mediated Fork Protection Synergizes with BRCA1 to Suppress Genomic Instability upon DNA Replication Stress.Mol Cell. 2018 Nov 1;72(3):568-582.e6. doi: 10.1016/j.molcel.2018.09.014. Epub 2018 Oct 18. Mol Cell. 2018. PMID: 30344097
-
Fanconi anemia proteins in telomere maintenance.DNA Repair (Amst). 2016 Jul;43:107-12. doi: 10.1016/j.dnarep.2016.02.007. Epub 2016 Apr 8. DNA Repair (Amst). 2016. PMID: 27118469 Free PMC article. Review.
-
CtIP: A DNA damage response protein at the intersection of DNA metabolism.DNA Repair (Amst). 2015 Aug;32:75-81. doi: 10.1016/j.dnarep.2015.04.016. Epub 2015 May 2. DNA Repair (Amst). 2015. PMID: 25957490 Review.
Cited by
-
A novel RBBP8(p.E281*) germline mutation is a predisposing mutation in familial hereditary cancer syndrome.J Mol Med (Berl). 2023 Oct;101(10):1255-1265. doi: 10.1007/s00109-023-02354-z. Epub 2023 Aug 24. J Mol Med (Berl). 2023. PMID: 37615686
-
Kinome-wide screening uncovers a role for Bromodomain Protein 3 in DNA double-stranded break repair.DNA Repair (Amst). 2023 Feb;122:103445. doi: 10.1016/j.dnarep.2022.103445. Epub 2022 Dec 24. DNA Repair (Amst). 2023. PMID: 36608404 Free PMC article.
-
To Join or Not to Join: Decision Points Along the Pathway to Double-Strand Break Repair vs. Chromosome End Protection.Front Cell Dev Biol. 2021 Jul 12;9:708763. doi: 10.3389/fcell.2021.708763. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34322492 Free PMC article. Review.
-
Homologous recombination within repetitive DNA.Curr Opin Genet Dev. 2021 Dec;71:143-153. doi: 10.1016/j.gde.2021.08.005. Epub 2021 Aug 28. Curr Opin Genet Dev. 2021. PMID: 34464817 Free PMC article. Review.
-
Telomere Maintenance and the cGAS-STING Pathway in Cancer.Cells. 2022 Jun 17;11(12):1958. doi: 10.3390/cells11121958. Cells. 2022. PMID: 35741087 Free PMC article. Review.
References
-
- de Lange T. Shelterin-mediated telomere protection. Annu. Rev. Genet. 2018; 52:223–247. - PubMed
-
- Griffith J.D., Comeau L., Rosenfield S., Stansel R.M., Bianchi A., Moss H., de Lange T.. Mammalian telomeres end in a large duplex loop. Cell. 1999; 97:503–514. - PubMed
-
- Harley C.B., Futcher A.B., Greider C.W.. Telomeres shorten during ageing of human fibroblasts. Nature. 1990; 345:458–460. - PubMed
-
- Baird D.M., Hendrickson E.A.. Telomeres and chromosomal translocations: there's a ligase at the end of the translocation. Adv. Exp. Med. Biol. 2018; 1044:89–112. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

