PKC induces release of a functional ectodomain of the guidance cue semaphorin6A
- PMID: 31378926
- PMCID: PMC6848763
- DOI: 10.1002/1873-3468.13561
PKC induces release of a functional ectodomain of the guidance cue semaphorin6A
Abstract
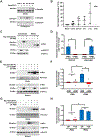
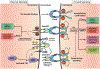
Semaphorins (Semas) are a family of secreted and transmembrane proteins that play critical roles in development. Interestingly, several vertebrate transmembrane Sema classes are capable of producing functional soluble ectodomains. However, little is known of soluble Sema6 ectodomains in the nervous system. Herein, we show that the soluble Sema6A ectodomain, sSema6A, exhibits natural and protein kinase C (PKC)-induced release. We show that PKC mediates Sema6A phosphorylation at specific sites and while this phosphorylation is not the primary mechanism regulating sSema6A production, we found that the intracellular domain confers resistance to ectodomain release. Finally, sSema6A is functional as it promotes the cohesion of zebrafish early eye field explants. This suggests that in addition to its canonical contact-mediated functions, Sema6A may have regulated, long-range, forward-signaling capacity.
Keywords: PKC; eye development; guidance cue; semaphorin; semaphorin6A; zebrafish.
© 2019 Federation of European Biochemical Societies.
Figures

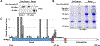


References
-
- Luo Y, Raible D & Raper JA (1993) Collapsin: a protein in brain that induces the collapse and paralysis of neuronal growth cones, Cell. 75, 217–27. - PubMed
-
- Kolodkin AL (1996) Semaphorins: mediators of repulsive growth cone guidance, Trends Cell Biol. 6, 15–22. - PubMed
-
- Jongbloets BC & Pasterkamp RJ (2014) Semaphorin signalling during development, Development. 141, 3292–7. - PubMed
-
- Kruger RP, Aurandt J & Guan KL (2005) Semaphorins command cells to move, Nat Rev Mol Cell Biol. 6, 789–800. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

