Half-Intercalation Stabilizes Slipped Mispairing and Explains Genome Vulnerability to Frameshift Mutagenesis by Endogenous "Molecular Bookmarks"
- PMID: 31379009
- PMCID: PMC6707839
- DOI: 10.1002/bies.201900062
Half-Intercalation Stabilizes Slipped Mispairing and Explains Genome Vulnerability to Frameshift Mutagenesis by Endogenous "Molecular Bookmarks"
Abstract
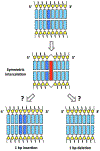
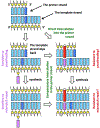
Some 60 years ago chemicals that intercalate between base pairs of duplex DNA were found to amplify frameshift mutagenesis. Surprisingly, the robust induction of frameshifts by intercalators still lacks a mechanistic model, leaving this classic phenomenon annoyingly intractable. A promising idea of asymmetric half-intercalation-stabilizing frameshift intermediates during DNA synthesis has never been developed into a model. Instead, researchers of frameshift mutagenesis embraced the powerful slipped-mispairing concept that unexpectedly struggled with the role of intercalators in frameshifting. It is proposed that the slipped mispairing and the half-intercalation ideas are two sides of the same coin. Further, existing findings are reviewed to test predictions of the combined "half-intercalation into the slipped-mispairing intermediate" model against accumulated knowledge. The existence of potential endogenous intercalators and the phenomenon of "DNA bookmarks" reveal ample possibilities for natural frameshift mutagenisis in the cell. From this alarming perspective, it is discussed how the cell could prevent genome deterioration from frameshift mutagenesis.
Keywords: 1 base pair (bp) indels; DNA synthesis; acridines; asymmetric intercalations; ethidium bromide; strand slippage.
© 2019 WILEY Periodicals, Inc.
Conflict of interest statement
The author declares no conflict of interest.
Figures





References
-
- Mirsky AE, Ris H, Nature 1949, 163, 666; H. Swift, Proc. Natl. Acad. Sci. U S A 1950, 36, 643; H. H. Swift, Physiol. Zool. 1950, 23, 169. - PubMed
-
- Franklin RE, Gosling RG, Nature 1953, 171, 740; R. E. Frankling, R. G. Gosling, Nature 1953, 172, 156. - PubMed
-
- Watson JD, Crick FHC, Nature 1953, 171, 737; J. D. Watson, F. H. C. Crick, Nature 1953, 171, 964. - PubMed
-
- Hollaender A, Annu. Rev. Physiol 1946, 8, 1. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

