Vineyard Soil Microbiome Composition Related to Rotundone Concentration in Australian Cool Climate 'Peppery' Shiraz Grapes
- PMID: 31379773
- PMCID: PMC6646731
- DOI: 10.3389/fmicb.2019.01607
Vineyard Soil Microbiome Composition Related to Rotundone Concentration in Australian Cool Climate 'Peppery' Shiraz Grapes
Abstract
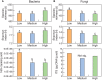
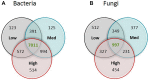
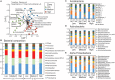
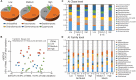
Soil microbial communities have an integral association with plants and play an important role in shaping plant nutrition, health, crop productivity and product quality. The influence of bacteria and fungi on wine fermentation is well known. However, little is known about the role of soil microbes, other than microbial pathogens, on grape composition or their role in vintage or site (terroir) impacts on grape composition. In this study, we used an amplicon sequencing approach to investigate the potential relationships between soil microbes and inherent spatial variation in grape metabolite composition - specifically, the concentration of the 'impact aroma compound' rotundone in Shiraz grapes (Vitis vinifera L.) grown in a 6.1 ha vineyard in the Grampians region of Victoria, Australia. Previous work had demonstrated temporal stability in patterns of within-vineyard spatial variation in rotundone concentration, enabling identification of defined 'zones' of inherently 'low' or 'high' concentration of this grape metabolite. 16S rRNA and ITS region-amplicon sequencing analysis of microbial communities in the surface soils collected from these zones indicated marked differences between zones in the genetic diversity and composition of the soil bacterial and fungal microbiome. Soils in the High rotundone zone exhibited higher diversity of bacteria, but lower diversity of fungi, compared to the soils in the Low rotundone zone. In addition, the network analysis of the microbial community in the High rotundone zone soils appeared well structured, especially with respect to the bacterial community, compared to that in the Low rotundone zone soils. The key differences in the microbial community structure between the rotundone zones are obvious for taxa/groups of both bacteria and fungi, particularly for bacteria belonging to Acidobacteria-GP4 and GP7, Rhizobiales, Gaiellaceae, Alphaproteobacteria and the Nectriaceae and Tremellaceae families of fungi. Although mulching in some parts of the vineyard caused changes in bacterial and fungal composition and overall microbial catabolic diversity and activity, its effects did not mask the rotundone zone-based variation. This finding of a systematic rotundone zone-based variation in soil microbiomes suggests an opportunity to bring together understanding of microbial ecology, plant biochemistry, and viticultural management for improved management of grape metabolism, composition and wine flavor.
Keywords: bacteria; fungi; grapes; microbiome diversity; rotundone.
Figures






References
-
- Anderson M. J. (2001). A new method for non-parametric multivariate analysis of variance. Austral. Ecol. 26 32–46. 10.1111/j.1442-9993.2001.01070.pp.x - DOI
-
- Backer R., Roken J. S., Ilangumaran G., Lamont J., Praslickova D., Ricci E., et al. (2018). Plant growth-promoting rhizobacteria: context, mechanisms of action, and roadmap to commercialization of biostimulants for sustainable agriculture. Front. Plant Sci. 9:1473. 10.3389/fpls.2018.01473 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

