Fecal and Mucosal Microbiota Profiling in Irritable Bowel Syndrome and Inflammatory Bowel Disease
- PMID: 31379797
- PMCID: PMC6650632
- DOI: 10.3389/fmicb.2019.01655
Fecal and Mucosal Microbiota Profiling in Irritable Bowel Syndrome and Inflammatory Bowel Disease
Abstract
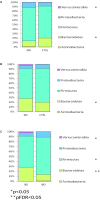
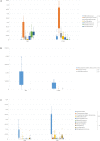
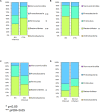
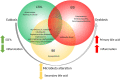
An imbalance in the bacterial species resulting in the loss of intestinal homeostasis has been described in inflammatory bowel diseases (IBD) and irritable bowel syndrome (IBS). In this prospective study, we investigated whether IBD and IBS patients exhibit specific changes in richness and distribution of fecal and mucosal-associated microbiota. Additionally, we assessed potential 16S rRNA gene amplicons biomarkers for IBD, IBS, and controls (CTRLs) by comparison of taxonomic composition. The relative abundance of bacteria, at phylum and genus/species levels, and the bacterial diversity were determined through 16S rRNA sequence-based fecal and mucosal microbiota analysis. Linear discriminant analysis effect size (LEfSe) was used for biomarker discovery associated to IBD and IBS as compared to CTRLs. In fecal and mucosal samples, the microbiota richness was characterized by a microbial diversity reduction, going from CTRLs to IBS to IBD. β-diversity analysis showed a clear separation between IBD and CTRLs and between IBD and IBS with no significant separation between IBS and CTRLs. β-diversity showed a clear separation between mucosa and stool samples in all the groups. In IBD, there was no difference between inflamed and not inflamed mucosa. Based upon the LEfSe data, the Anaerostipes and Ruminococcaceae were identified as the most differentially abundant bacterial taxa in CTRLs. Erysipelotrichi was identified as potential biomarker for IBS, while Gammaproteobacteria, Enterococcus, and Enterococcaceae for IBD. This study provides an overview of the alterations of microbiota and may aid in identifying potential 16S rRNA gene amplicons mucosal biomarkers for IBD and IBS.
Keywords: dysbiosis; gut microbiota; inflammation; inflammatory bowel disease; irritable bowel syndrome.
Figures

References
-
- Baumgart M., Dogan B., Rishniw M., Weitzman G., Bosworth B., Yantiss R., et al. (2007). Culture independent analysis of ileal mucosa reveals a selective increase in invasive Escherichia coli of novel phylogeny relative to depletion of Clostridiales in Crohn’s disease involving the ileum. ISME J. 1 403–418. 10.1038/ismej.2007.52 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

