Genome Analysis Reveals Genetic Admixture and Signature of Selection for Productivity and Environmental Traits in Iraqi Cattle
- PMID: 31379916
- PMCID: PMC6646475
- DOI: 10.3389/fgene.2019.00609
Genome Analysis Reveals Genetic Admixture and Signature of Selection for Productivity and Environmental Traits in Iraqi Cattle
Abstract
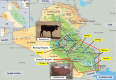
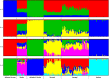
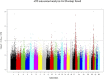
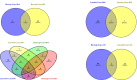
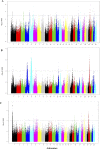
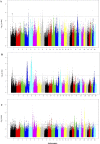
The Near East cattle are adapted to different agro-ecological zones including desert areas, mountains habitats, and humid regions along the Tigris and Euphrates rivers system. The region was one of the earliest and most significant areas of cattle husbandry. Currently, four main breeds of Iraqi cattle are recognized. Among these, the Jenoubi is found in the southern more humid part of Iraq, while the Rustaqi is found in the middle and drier region of the country. Despite their importance, Iraqi cattle have up to now been poorly characterized at the genome level. Here, we report at a genome-wide level the diversity and signature of positive selection in these two breeds. Thirty-five unrelated Jenoubi cattle, sampled in the Maysan and Basra regions, and 60 Rustaqi cattle, from around Baghdad and Babylon, were genotyped using the Illumina Bovine HD BeadChip (700K). Genetic population structure and diversity level were studied using principal component analysis (PCA), expected heterozygosity (He), observed heterozygosity (Ho), and admixture. Signatures of selection were studied using extended haplotype homozygosity (EHH) (iHS and Rsb) and inter-population Wright's Fst. The results of PCA and admixture analysis, including European taurine, Asian indicine, African indicine, and taurine indicate that the two breeds are crossbreed zebu × taurine, with more zebu background in Jenoubi cattle compared with Rustaqi. The Rustaqi has the greatest mean heterozygosity (He = 0.37) among all breeds. iHS and Rsb signatures of selection analyses identify 68 candidate genes under positive selection in the two Iraqi breeds, while Fst analysis identifies 220 candidate genes including genes related to the innate and acquired immunity responses, different environmental selection pressures (e.g., tick resistance and heat stress), and genes of commercial interest (e.g., marbling score).
Keywords: Bos indicus; Bos taurus; adaptive genes; diversity; genetic structure; immune responses; positive selection.
Figures




References
-
- Al-Bayatti S., Essa A., Nasef N., Salman S., Abdulkader N., Al-Tameemi R., Mahdi L. (2016). Phenotypic characterisation of Rustaqi cattle breed within its natural environment. Proceeding of Regional Conference for Animal Genetic Resources Conservation, Oman, pp. 22.
-
- Al-Murrani W., Majid S., Alkas J. (2003). Animal genetic resources in Iraq. Country scientific report. Baghdad: Iraq Agricultural Ministry Publication, 1–55.
-
- Al-Ramahi H., Kshash Q. (2011). Epidemiological study of some factors affecting prevalence of clinical theileriosis in cows in southern Baghdad. Euphrates J. Agric. Sci. 3 (3), 22–28.
LinkOut - more resources
Full Text Sources
Miscellaneous

