MIG1 Glucose Repression in Metabolic Processes of Saccharomyces cerevisiae: Genetics to Metabolic Engineering
- PMID: 31379993
- PMCID: PMC6626512
MIG1 Glucose Repression in Metabolic Processes of Saccharomyces cerevisiae: Genetics to Metabolic Engineering
Abstract
Background: Although Saccharomyces cerevisiae has several industrial applications, there are still fundamental problems associated with sequential use of carbon sources. As such, glucose repression effect can direct metabolism of yeast to preferably anaerobic conditions. This leads to higher ethanol production and less efficient production of recombinant products. The general glucose repression system is constituted by MIG1, TUP1 and SSN6 factors. The role of MIG1 is known in glucose repression but the evaluation of effects on aerobic/anaerobic metabolism by deletion of MIG1 and constructing an optimal strain brand remains unclear and an objective to be explored.
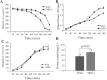
Methods: To find the impact of MIG1 in induction of glucose-repression, the Mig1 disruptant strain (ΔMIG1) was produced for comparing with its congenic wild-type strain (2805). The analysis approached for changes in the rate of glucose consumption, biomass yield, cell protein contents, ethanol and intermediate metabolites production. The MIG1 disruptant strain exhibited 25% glucose utilization, 12% biomass growth rate and 22% protein content over the wild type. The shift to respiratory pathway has been demonstrated by 122.86 and 40% increase of glycerol and pyruvate production, respectively as oxidative metabolites, while the reduction of fermentative metabolites such as acetate 35.48 and ethanol 24%.
Results: Results suggest that ΔMIG1 compared to the wild-type strain can significantly present less effects of glucose repression.
Conclusion: The constructed strain has more efficient growth in aerobic cultivations and it can be a potential host for biotechnological recombinant yields and industrial interests.
Keywords: Homologous recombination; Metabolic pathways; Saccharomyces cerevisiae; Yeasts.
Figures


Similar articles
-
Saccharomyces cerevisiae, key role of MIG1 gene in metabolic switching: putative fermentation/oxidation.J Biol Regul Homeost Agents. 2018 May-Jun;32(3):649-654. J Biol Regul Homeost Agents. 2018. PMID: 29921394
-
Improving Xylose Utilization of Saccharomyces cerevisiae by Expressing the MIG1 Mutant from the Self-Flocculating Yeast SPSC01.Protein Pept Lett. 2018;25(2):202-207. doi: 10.2174/0929866525666180122142609. Protein Pept Lett. 2018. PMID: 29359658
-
Effects of MIG1, TUP1 and SSN6 deletion on maltose metabolism and leavening ability of baker's yeast in lean dough.Microb Cell Fact. 2014 Jul 4;13:93. doi: 10.1186/s12934-014-0093-4. Microb Cell Fact. 2014. PMID: 24993311 Free PMC article.
-
Production of fuels and chemicals from xylose by engineered Saccharomyces cerevisiae: a review and perspective.Microb Cell Fact. 2017 May 11;16(1):82. doi: 10.1186/s12934-017-0694-9. Microb Cell Fact. 2017. PMID: 28494761 Free PMC article. Review.
-
Rewiring yeast metabolism to synthesize products beyond ethanol.Curr Opin Chem Biol. 2020 Dec;59:182-192. doi: 10.1016/j.cbpa.2020.08.005. Epub 2020 Oct 5. Curr Opin Chem Biol. 2020. PMID: 33032255 Free PMC article. Review.
Cited by
-
Past, Present, and Future Perspectives on Whey as a Promising Feedstock for Bioethanol Production by Yeast.J Fungi (Basel). 2022 Apr 12;8(4):395. doi: 10.3390/jof8040395. J Fungi (Basel). 2022. PMID: 35448626 Free PMC article. Review.
-
Convergence between Regulation of Carbon Utilization and Catabolic Repression in Xanthophyllomyces dendrorhous.mSphere. 2020 Apr 1;5(2):e00065-20. doi: 10.1128/mSphere.00065-20. mSphere. 2020. PMID: 32238568 Free PMC article.
-
Effects of the carbon source on the physiology and invertase activity of the yeast Saccharomyces cerevisiae FT858.3 Biotech. 2020 Aug;10(8):348. doi: 10.1007/s13205-020-02335-w. Epub 2020 Jul 22. 3 Biotech. 2020. PMID: 32728515 Free PMC article.
References
-
- Xu JR, Zhao XQ, Liu CG, Bai FW. Improving xylose utilization of Saccharomyces cerevisiae by expressing the MIG1 mutant from the self-flocculating yeast SPSC 01. Protein Pept Lett 2018;25(2):202–207. - PubMed
-
- Galello F, Pautasso C, Reca S, Cañonero L, Portela P, Moreno S, et al. Transcriptional regulation of the protein kinase a subunits in Saccharomyces cerevisiae during fermentative growth. Yeast 2017;34(12):495–508. - PubMed
-
- Klein CJL, Rasmussen JJ, Rønnow B, Olsson L, Nielsen J. Investigation of the impact of MIG1 and MIG2 on the physiology of Saccharomyces cerevisiae. J Biotech 1999;68(2–3):197–212. - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
