MftD Catalyzes the Formation of a Biologically Active Redox Center in the Biosynthesis of the Ribosomally Synthesized and Post-translationally Modified Redox Cofactor Mycofactocin
- PMID: 31381312
- PMCID: PMC6716157
- DOI: 10.1021/jacs.9b06102
MftD Catalyzes the Formation of a Biologically Active Redox Center in the Biosynthesis of the Ribosomally Synthesized and Post-translationally Modified Redox Cofactor Mycofactocin
Abstract
Mycofactocin (MFT) is a putative ribosomally synthesized and post-translationally modified (RiPP) redox cofactor. The biosynthesis of MFT is encoded by the gene cluster mftABCDEF. While processing of the precursor peptide by MftB, MftC, and MftE has been shown to result in the formation of the small molecule 3-amino-5-[(p-hydroxyphenyl)methyl]-4,4-dimethyl-2-pyrrolidinone (AHDP), no activity has been shown for the putative dehydrogenase MftD and the putative glycosyltransferase MftF. In addition, evidence demonstrating that MFT is a redox cofactor has only been limited to the requirement of mft genes for ethanol assimilation in Mycobacterium smegmatis mc2155. Here, we demonstrate that MftD catalyzes the oxidative deamination of AHDP, forming an α-keto moiety on the resulting molecule, which we call pre-mycofactocin (PMFT). We characterize PMFT by 1D and 2D NMR spectroscopy techniques and by high-resolution mass spectrometry data to solve its structure. We further characterized PMFT by cyclic voltammetry and found its midpoint potential to be ∼255 mV. Lastly, we demonstrate that PMFT is a biologically active redox cofactor that oxidizes NADH bound by M. smegmatis carveol dehydrogenase (MsCDH) and can be used by MsCDH in the oxidation of carveol. These data demonstrate for the first time that PMFT functions as a biologically active redox mediator and provides the most direct evidence to date that MFT is a RiPP-derived redox cofactor.
Figures




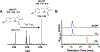



Similar articles
-
Mycofactocin Is Associated with Ethanol Metabolism in Mycobacteria.mBio. 2019 May 21;10(3):e00190-19. doi: 10.1128/mBio.00190-19. mBio. 2019. PMID: 31113891 Free PMC article.
-
Occurrence, function, and biosynthesis of mycofactocin.Appl Microbiol Biotechnol. 2019 Apr;103(7):2903-2912. doi: 10.1007/s00253-019-09684-4. Epub 2019 Feb 18. Appl Microbiol Biotechnol. 2019. PMID: 30778644 Free PMC article. Review.
-
The Creatininase Homolog MftE from Mycobacterium smegmatis Catalyzes a Peptide Cleavage Reaction in the Biosynthesis of a Novel Ribosomally Synthesized Post-translationally Modified Peptide (RiPP).J Biol Chem. 2017 Mar 10;292(10):4371-4381. doi: 10.1074/jbc.M116.762062. Epub 2017 Jan 11. J Biol Chem. 2017. PMID: 28077628 Free PMC article.
-
Structure elucidation of the redox cofactor mycofactocin reveals oligo-glycosylation by MftF.Chem Sci. 2020 Apr 23;11(20):5182-5190. doi: 10.1039/d0sc01172j. eCollection 2020 May 28. Chem Sci. 2020. PMID: 33014324 Free PMC article.
-
Recent insights into the biosynthesis and biological activities of the peptide-derived redox cofactor mycofactocin.Nat Prod Rep. 2025 Aug 13;42(8):1344-1366. doi: 10.1039/d5np00012b. Nat Prod Rep. 2025. PMID: 40375824 Review.
Cited by
-
The manifold roles of microbial ribosomal peptide-based natural products in physiology and ecology.J Biol Chem. 2020 Jan 3;295(1):34-54. doi: 10.1074/jbc.REV119.006545. Epub 2019 Nov 29. J Biol Chem. 2020. PMID: 31784450 Free PMC article. Review.
-
Bicyclostreptins are radical SAM enzyme-modified peptides with unique cyclization motifs.Nat Chem Biol. 2022 Oct;18(10):1135-1143. doi: 10.1038/s41589-022-01090-8. Epub 2022 Aug 11. Nat Chem Biol. 2022. PMID: 35953547
-
Role of Premycofactocin Synthase in Growth, Microaerophilic Adaptation, and Metabolism of Mycobacterium tuberculosis.mBio. 2021 Aug 31;12(4):e0166521. doi: 10.1128/mBio.01665-21. Epub 2021 Jul 27. mBio. 2021. PMID: 34311585 Free PMC article.
-
A mycofactocin-associated dehydrogenase is essential for ethylene glycol metabolism by Rhodococcus jostii RHA1.Appl Microbiol Biotechnol. 2024 Dec;108(1):58. doi: 10.1007/s00253-023-12966-7. Epub 2024 Jan 4. Appl Microbiol Biotechnol. 2024. PMID: 38175243
-
Metabolic profiling of dormant Mycolicibacterium smegmatis cells' reactivation reveals a gradual assembly of metabolic processes.Metabolomics. 2020 Feb 6;16(2):24. doi: 10.1007/s11306-020-1645-8. Metabolomics. 2020. PMID: 32025943
References
-
- Arnison PG; Bibb MJ; Bierbaum G; Bowers AA; Bugni TS; Bulaj G; Camarero JA; Campopiano DJ; Challis GL; Clardy J; Cotter PD; Craik DJ; Dawson M; Dittmann E; Donadio S; Dorrestein PC.; Entian K-D; Fischbach MA; Garavelli JS; Göransson U; Gruber CW; Haft DH; Hemscheidt TK; Hertweck C; Hill C; Horswill AR; Jaspars M; Kelly WL; Klinman JP; Kuipers OP; Link AJ; Liu W; Marahiel MA; Mitchell DA; Moll GN; Moore BS; Müller R; Nair SK; Nes IF; Norris GE; Olivera BM; Onaka H; Patchett ML; Piel J; Reaney MJT; Rebuffat S; Ross RP; Sahl H-G; Schmidt EW; Selsted ME; Severinov K; Shen B; Sivonen K; Smith L; Stein T; Süssmuth RD; Tagg JR; Tang G-L; Truman AW; Vederas JC; Walsh CT; Walton JD; Wenzel SC; Willey JM; van der Donk WA. Ribosomally Synthesized and Post-Translationally Modified Peptide Natural Products: Overview and Recommendations for a Universal Nomenclature. Nat. Prod. Rep 2013, 30 (1), 108–160. 10.1039/C2NP20085F. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

