Methyl-CpG-binding domain 9 (MBD9) is required for H2A.Z incorporation into chromatin at a subset of H2A.Z-enriched regions in the Arabidopsis genome
- PMID: 31381567
- PMCID: PMC6695207
- DOI: 10.1371/journal.pgen.1008326
Methyl-CpG-binding domain 9 (MBD9) is required for H2A.Z incorporation into chromatin at a subset of H2A.Z-enriched regions in the Arabidopsis genome
Abstract
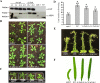
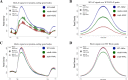
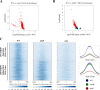
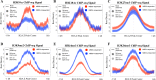
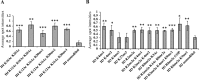
The SWR1 chromatin remodeling complex, which deposits the histone variant H2A.Z into nucleosomes, has been well characterized in yeast and animals, but its composition in plants has remained uncertain. We used the conserved SWR1 subunit ACTIN RELATED PROTEIN 6 (ARP6) as bait in tandem affinity purification experiments to isolate associated proteins from Arabidopsis thaliana. We identified all 11 subunits found in yeast SWR1 and the homologous mammalian SRCAP complexes, demonstrating that this complex is conserved in plants. We also identified several additional proteins not previously associated with SWR1, including Methyl-CpG-BINDING DOMAIN 9 (MBD9) and three members of the Alfin1-like protein family, all of which have been shown to bind modified histone tails. Since mbd9 mutant plants were phenotypically similar to arp6 mutants, we explored a potential role for MBD9 in H2A.Z deposition. We found that MBD9 is required for proper H2A.Z incorporation at thousands of discrete sites, which represent a subset of the genomic regions normally enriched with H2A.Z. We also discovered that MBD9 preferentially interacts with acetylated histone H4 peptides, as well as those carrying mono- or dimethylated H3 lysine 4, or dimethylated H3 arginine 2 or 8. Considering that MBD9-dependent H2A.Z sites show a distinct histone modification profile, we propose that MBD9 recognizes particular nucleosome modifications via its PHD- and Bromo-domains and thereby guides SWR1 to these sites for H2A.Z deposition. Our data establish the SWR1 complex as being conserved across eukaryotes and suggest that MBD9 may be involved in targeting the complex to specific genomic sites through nucleosomal interactions. The finding that MBD9 does not appear to be a core subunit of the Arabidopsis SWR1 complex, along with the synergistic phenotype of arp6;mbd9 double mutants, suggests that MBD9 also has important roles beyond H2A.Z deposition.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


Similar articles
-
Arabidopsis SWR1-associated protein methyl-CpG-binding domain 9 is required for histone H2A.Z deposition.Nat Commun. 2019 Jul 26;10(1):3352. doi: 10.1038/s41467-019-11291-w. Nat Commun. 2019. PMID: 31350403 Free PMC article.
-
A plant-specific SWR1 chromatin-remodeling complex couples histone H2A.Z deposition with nucleosome sliding.EMBO J. 2020 Apr 1;39(7):e102008. doi: 10.15252/embj.2019102008. Epub 2020 Mar 2. EMBO J. 2020. PMID: 32115743 Free PMC article.
-
NAP1-RELATED PROTEIN1 and 2 negatively regulate H2A.Z abundance in chromatin in Arabidopsis.Nat Commun. 2020 Jun 8;11(1):2887. doi: 10.1038/s41467-020-16691-x. Nat Commun. 2020. PMID: 32513971 Free PMC article.
-
SWR1 Chromatin Remodeling Complex: A Key Transcriptional Regulator in Plants.Cells. 2019 Dec 12;8(12):1621. doi: 10.3390/cells8121621. Cells. 2019. PMID: 31842357 Free PMC article. Review.
-
Roles of the INO80 and SWR1 Chromatin Remodeling Complexes in Plants.Int J Mol Sci. 2019 Sep 17;20(18):4591. doi: 10.3390/ijms20184591. Int J Mol Sci. 2019. PMID: 31533258 Free PMC article. Review.
Cited by
-
Replacement of Arabidopsis H2A.Z with human H2A.Z orthologs reveals extensive functional conservation and limited importance of the N-terminal tail sequence for Arabidopsis development.bioRxiv [Preprint]. 2024 Jul 12:2023.11.03.565555. doi: 10.1101/2023.11.03.565555. bioRxiv. 2024. Update in: Genetics. 2025 Jun 4;230(2):iyaf065. doi: 10.1093/genetics/iyaf065. PMID: 37961174 Free PMC article. Updated. Preprint.
-
Interactome of Arabidopsis Thaliana.Plants (Basel). 2022 Jan 27;11(3):350. doi: 10.3390/plants11030350. Plants (Basel). 2022. PMID: 35161331 Free PMC article.
-
Spatiotemporal control of miR398 biogenesis, via chromatin remodeling and kinase signaling, ensures proper ovule development.Plant Cell. 2021 Jul 2;33(5):1530-1553. doi: 10.1093/plcell/koab056. Plant Cell. 2021. PMID: 33570655 Free PMC article.
-
Molecular mechanisms and biological functions of active DNA demethylation in plants.Epigenetics Chromatin. 2025 Jul 5;18(1):41. doi: 10.1186/s13072-025-00605-6. Epigenetics Chromatin. 2025. PMID: 40618161 Free PMC article. Review.
-
Contribution of the histone variant H2A.Z to expression of responsive genes in plants.Semin Cell Dev Biol. 2023 Feb 15;135:85-92. doi: 10.1016/j.semcdb.2022.04.006. Epub 2022 Apr 23. Semin Cell Dev Biol. 2023. PMID: 35474148 Free PMC article. Review.
References
-
- Mizuguchi G, Shen X, Landry J, Wu W, Sen S, Wu C. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science (New York, NY). 2004;303(5656):343–8. - PubMed
-
- Jarillo J, Pineiro M. H2A.Z mediates different aspects of chromatin function and modulates flowering responses in Arabidopsis. The Plant journal: for cell and molecular biology. 2015;83(1):96–109. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

