Bipartite interface of the measles virus phosphoprotein X domain with the large polymerase protein regulates viral polymerase dynamics
- PMID: 31381607
- PMCID: PMC6695210
- DOI: 10.1371/journal.ppat.1007995
Bipartite interface of the measles virus phosphoprotein X domain with the large polymerase protein regulates viral polymerase dynamics
Abstract
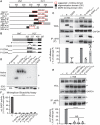
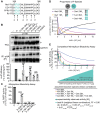
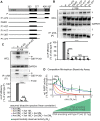
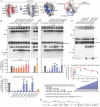
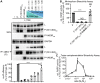
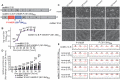
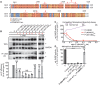
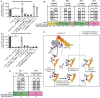
Measles virus (MeV) is a highly contagious, re-emerging, major human pathogen. Replication requires a viral RNA-dependent RNA polymerase (RdRP) consisting of the large (L) polymerase protein complexed with the homo-tetrameric phosphoprotein (P). In addition, P mediates interaction with the nucleoprotein (N)-encapsidated viral RNA genome. The nature of the P:L interface and RdRP negotiation of the ribonucleoprotein template are poorly understood. Based on biochemical interface mapping, swapping of the central P tetramerization domain (OD) for yeast GCN4, and functional assays, we demonstrate that the MeV P-to-L interface is bipartite, comprising a coiled-coil microdomain proximal to the OD and an unoccupied face of the triangular prism-shaped C-terminal P X-domain (P-XD), which is distinct from the known P-XD face that binds N-tail. Mixed null-mutant P tetramers regained L-binding competence in a ratio-dependent manner and fully reclaimed bioactivity in minireplicon assays and recombinant MeV, demonstrating that the individual L-binding interface elements are physically and mechanistically distinct. P-XD binding competence to L and N was likewise trans-complementable, which, combined with mathematical modeling, enabled the mechanistic characterization of P through two- and stoichiometrically-controlled three-way complementations. Only one each of the four XDs per P tetramer must be L or N binding-competent for bioactivity, but interaction of the same P-XD with L and N was mutually exclusive, and L binding superseded engaging N. Mixed P tetramers with a single, designated L binding-competent P-XD caused significant RdRP hyperactivity, outlining a model of iterative resolution and reformation of the P-XD:L interface regulating polymerase mobility.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
Similar articles
-
A three-way interface of the Nipah virus phosphoprotein X-domain coordinates polymerase movement along the viral genome.J Virol. 2024 Oct 22;98(10):e0098624. doi: 10.1128/jvi.00986-24. Epub 2024 Sep 4. J Virol. 2024. PMID: 39230304 Free PMC article.
-
Viral evolution identifies a regulatory interface between paramyxovirus polymerase complex and nucleocapsid that controls replication dynamics.Sci Adv. 2020 Mar 4;6(10):eaaz1590. doi: 10.1126/sciadv.aaz1590. eCollection 2020 Mar. Sci Adv. 2020. PMID: 32181359 Free PMC article.
-
Plasticity in structural and functional interactions between the phosphoprotein and nucleoprotein of measles virus.J Biol Chem. 2012 Apr 6;287(15):11951-67. doi: 10.1074/jbc.M111.333088. Epub 2012 Feb 8. J Biol Chem. 2012. PMID: 22318731 Free PMC article.
-
Modulation of Measles Virus NTAIL Interactions through Fuzziness and Sequence Features of Disordered Binding Sites.Biomolecules. 2018 Dec 27;9(1):8. doi: 10.3390/biom9010008. Biomolecules. 2018. PMID: 30591682 Free PMC article. Review.
-
The measles virus N(TAIL)-XD complex: an illustrative example of fuzziness.Adv Exp Med Biol. 2012;725:126-41. doi: 10.1007/978-1-4614-0659-4_8. Adv Exp Med Biol. 2012. PMID: 22399322 Review.
Cited by
-
Structures of the mumps virus polymerase complex via cryo-electron microscopy.Nat Commun. 2024 May 17;15(1):4189. doi: 10.1038/s41467-024-48389-9. Nat Commun. 2024. PMID: 38760379 Free PMC article.
-
The Nucleocapsid of Paramyxoviruses: Structure and Function of an Encapsidated Template.Viruses. 2021 Dec 9;13(12):2465. doi: 10.3390/v13122465. Viruses. 2021. PMID: 34960734 Free PMC article. Review.
-
Transient Non-local Interactions Dominate the Dynamics of Measles Virus NTAIL.bioRxiv [Preprint]. 2025 Jun 17:2024.07.22.604679. doi: 10.1101/2024.07.22.604679. bioRxiv. 2025. PMID: 39091801 Free PMC article. Preprint.
-
Evolutionary history of cotranscriptional editing in the paramyxoviral phosphoprotein gene.Virus Evol. 2021 Mar 27;7(1):veab028. doi: 10.1093/ve/veab028. eCollection 2021 Jan. Virus Evol. 2021. PMID: 34141448 Free PMC article. Review.
-
The C Protein Is Recruited to Measles Virus Ribonucleocapsids by the Phosphoprotein.J Virol. 2020 Jan 31;94(4):e01733-19. doi: 10.1128/JVI.01733-19. Print 2020 Jan 31. J Virol. 2020. PMID: 31748390 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

