The alpha-1 subunit of the Na+,K+-ATPase (ATP1A1) is required for macropinocytic entry of respiratory syncytial virus (RSV) in human respiratory epithelial cells
- PMID: 31381610
- PMCID: PMC6695199
- DOI: 10.1371/journal.ppat.1007963
The alpha-1 subunit of the Na+,K+-ATPase (ATP1A1) is required for macropinocytic entry of respiratory syncytial virus (RSV) in human respiratory epithelial cells
Abstract
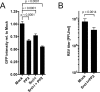
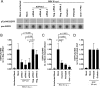
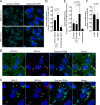
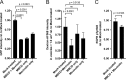
Human respiratory syncytial virus (RSV) is the leading viral cause of acute pediatric lower respiratory tract infections worldwide, with no available vaccine or effective antiviral drug. To gain insight into virus-host interactions, we performed a genome-wide siRNA screen. The expression of over 20,000 cellular genes was individually knocked down in human airway epithelial A549 cells, followed by infection with RSV expressing green fluorescent protein (GFP). Knockdown of expression of the cellular ATP1A1 protein, which is the major subunit of the Na+,K+-ATPase of the plasma membrane, had one of the strongest inhibitory effects on GFP expression and viral titer. Inhibition was not observed for vesicular stomatitis virus, indicating that it was RSV-specific rather than a general effect. ATP1A1 formed clusters in the plasma membrane very early following RSV infection, which was independent of replication but dependent on the attachment glycoprotein G. RSV also triggered activation of ATP1A1, resulting in signaling by c-Src-kinase activity that transactivated epidermal growth factor receptor (EGFR) by Tyr845 phosphorylation. ATP1A1 signaling and activation of both c-Src and EGFR were found to be required for efficient RSV uptake. Signaling events downstream of EGFR culminated in the formation of macropinosomes. There was extensive uptake of RSV virions into macropinosomes at the beginning of infection, suggesting that this is a major route of RSV uptake, with fusion presumably occurring in the macropinosomes rather than at the plasma membrane. Important findings were validated in primary human small airway epithelial cells (HSAEC). In A549 cells and HSAEC, RSV uptake could be inhibited by the cardiotonic steroid ouabain and the digitoxigenin derivative PST2238 (rostafuroxin) that bind specifically to the ATP1A1 extracellular domain and block RSV-triggered EGFR Tyr845 phosphorylation. In conclusion, we identified ATP1A1 as a host protein essential for macropinocytic entry of RSV into respiratory epithelial cells, and identified PST2238 as a potential anti-RSV drug.
Conflict of interest statement
M.L., P.L.C. and S.M. are coinventors on a patent application for the development of PST2238 (rostafuroxin) as an antiviral drug.
Figures









Similar articles
-
ATP1A1-mediated Src signaling inhibits coronavirus entry into host cells.J Virol. 2015 Apr;89(8):4434-48. doi: 10.1128/JVI.03274-14. Epub 2015 Feb 4. J Virol. 2015. PMID: 25653449 Free PMC article.
-
Targeting Intracellular Ion Homeostasis for the Control of Respiratory Syncytial Virus.Am J Respir Cell Mol Biol. 2018 Dec;59(6):733-744. doi: 10.1165/rcmb.2017-0345OC. Am J Respir Cell Mol Biol. 2018. PMID: 30095982
-
Host cell entry of respiratory syncytial virus involves macropinocytosis followed by proteolytic activation of the F protein.PLoS Pathog. 2013;9(4):e1003309. doi: 10.1371/journal.ppat.1003309. Epub 2013 Apr 11. PLoS Pathog. 2013. PMID: 23593008 Free PMC article.
-
New insights on the viral and host factors contributing to the airway pathogenesis caused by the respiratory syncytial virus.Crit Rev Microbiol. 2016 Sep;42(5):800-12. doi: 10.3109/1040841X.2015.1055711. Epub 2015 Jun 29. Crit Rev Microbiol. 2016. PMID: 26119025 Review.
-
Binding and entry of respiratory syncytial virus into host cells and initiation of the innate immune response.Cell Microbiol. 2003 Oct;5(10):671-80. doi: 10.1046/j.1462-5822.2003.00313.x. Cell Microbiol. 2003. PMID: 12969373 Review.
Cited by
-
Advances in development of antiviral strategies against respiratory syncytial virus.Acta Pharm Sin B. 2025 Apr;15(4):1752-1772. doi: 10.1016/j.apsb.2025.02.015. Epub 2025 Feb 20. Acta Pharm Sin B. 2025. PMID: 40486844 Free PMC article. Review.
-
PAR-2-activated secretion by airway gland serous cells: role for CFTR and inhibition by Pseudomonas aeruginosa.Am J Physiol Lung Cell Mol Physiol. 2021 May 1;320(5):L845-L879. doi: 10.1152/ajplung.00411.2020. Epub 2021 Mar 3. Am J Physiol Lung Cell Mol Physiol. 2021. PMID: 33655758 Free PMC article.
-
Respiratory syncytial virus fuses with plasma membrane to infect primary cultures of bronchial epithelial cells.Front Microbiol. 2025 Feb 26;16:1498955. doi: 10.3389/fmicb.2025.1498955. eCollection 2025. Front Microbiol. 2025. PMID: 40099186 Free PMC article.
-
Cardiac glycosides: Looking beyond heart failure and atrial fibrillation.Indian J Pharmacol. 2025 Jan 1;57(1):33-47. doi: 10.4103/ijp.ijp_934_24. Epub 2025 May 6. Indian J Pharmacol. 2025. PMID: 40324829 Free PMC article.
-
Na+/K+-ATPase as a Target of Cardiac Glycosides for the Treatment of SARS-CoV-2 Infection.Front Pharmacol. 2021 Apr 15;12:624704. doi: 10.3389/fphar.2021.624704. eCollection 2021. Front Pharmacol. 2021. PMID: 33935717 Free PMC article. Review.
References
-
- Lozano R, Naghavi M, Foreman K, Lim S, Shibuya K, Aboyans V, et al. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet. 2012;380(9859):2095–128. Epub 2012/12/19. 10.1016/S0140-6736(12)61728-0 . - DOI - PMC - PubMed
-
- Shi T, McAllister DA, O'Brien KL, Simoes EAF, Madhi SA, Gessner BD, et al. Global, regional, and national disease burden estimates of acute lower respiratory infections due to respiratory syncytial virus in young children in 2015: a systematic review and modelling study. Lancet. 2017;390(10098):946–58. Epub 2017/07/12. 10.1016/S0140-6736(17)30938-8 - DOI - PMC - PubMed
-
- Collins PL, Karron RA. Respiratory Syncytial Virus and Metapneumovirus In: Knipe DM, Howley PM, Cohen JI, Griffin DE, Lamb RA, Martin MA, et al., editors. Fields virology. 1 6th ed Philadelphia: Lippincott Williams&Wilkins; 2013. p. 1086–123.
-
- Bukreyev A, Whitehead SS, Murphy BR, Collins PL. Recombinant respiratory syncytial virus from which the entire SH gene has been deleted grows efficiently in cell culture and exhibits site-specific attenuation in the respiratory tract of the mouse. J Virol. 1997;71(12):8973–82. ; PubMed Central PMCID: 230197. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

