Molecular Interactions Between Reactive Oxygen Species and Autophagy in Kidney Disease
- PMID: 31382550
- PMCID: PMC6696055
- DOI: 10.3390/ijms20153791
Molecular Interactions Between Reactive Oxygen Species and Autophagy in Kidney Disease
Abstract
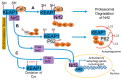
Reactive oxygen species (ROS) are highly reactive signaling molecules that maintain redox homeostasis in mammalian cells. Dysregulation of redox homeostasis under pathological conditions results in excessive generation of ROS, culminating in oxidative stress and the associated oxidative damage of cellular components. ROS and oxidative stress play a vital role in the pathogenesis of acute kidney injury and chronic kidney disease, and it is well documented that increased oxidative stress in patients enhances the progression of renal diseases. Oxidative stress activates autophagy, which facilitates cellular adaptation and diminishes oxidative damage by degrading and recycling intracellular oxidized and damaged macromolecules and dysfunctional organelles. In this review, we report the current understanding of the molecular regulation of autophagy in response to oxidative stress in general and in the pathogenesis of kidney diseases. We summarize how the molecular interactions between ROS and autophagy involve ROS-mediated activation of autophagy and autophagy-mediated reduction of oxidative stress. In particular, we describe how ROS impact various signaling pathways of autophagy, including mTORC1-ULK1, AMPK-mTORC1-ULK1, and Keap1-Nrf2-p62, as well as selective autophagy including mitophagy and pexophagy. Precise elucidation of the molecular mechanisms of interactions between ROS and autophagy in the pathogenesis of renal diseases may identify novel targets for development of drugs for preventing renal injury.
Keywords: AMPK; acute kidney injury; autophagy; chronic kidney disease; diabetic nephropathy; mTORC1; oxidants; reactive oxygen species.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

