Metabolic flux analysis for metabolome data validation of naturally xylose-fermenting yeasts
- PMID: 31382948
- PMCID: PMC6683545
- DOI: 10.1186/s12896-019-0548-0
Metabolic flux analysis for metabolome data validation of naturally xylose-fermenting yeasts
Abstract
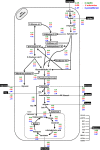
Background: Efficient xylose fermentation still demands knowledge regarding xylose catabolism. In this study, metabolic flux analysis (MFA) and metabolomics were used to improve our understanding of xylose metabolism. Thus, a stoichiometric model was constructed to simulate the intracellular carbon flux and used to validate the metabolome data collected within xylose catabolic pathways of non-Saccharomyces xylose utilizing yeasts.
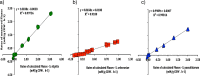
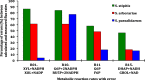
Results: A metabolic flux model was constructed using xylose fermentation data from yeasts Scheffersomyces stipitis, Spathaspora arborariae, and Spathaspora passalidarum. In total, 39 intracellular metabolic reactions rates were utilized validating the measurements of 11 intracellular metabolites, acquired by mass spectrometry. Among them, 80% of total metabolites were confirmed with a correlation above 90% when compared to the stoichiometric model. Among the intracellular metabolites, fructose-6-phosphate, glucose-6-phosphate, ribulose-5-phosphate, and malate are validated in the three studied yeasts. However, the metabolites phosphoenolpyruvate and pyruvate could not be confirmed in any yeast. Finally, the three yeasts had the metabolic fluxes from xylose to ethanol compared. Xylose catabolism occurs at twice-higher flux rates in S. stipitis than S. passalidarum and S. arborariae. Besides, S. passalidarum present 1.5 times high flux rate in the xylose reductase reaction NADH-dependent than other two yeasts.
Conclusions: This study demonstrated a novel strategy for metabolome data validation and brought insights about naturally xylose-fermenting yeasts. S. stipitis and S. passalidarum showed respectively three and twice higher flux rates of XR with NADH cofactor, reducing the xylitol production when compared to S. arborariae. Besides then, the higher flux rates directed to pentose phosphate pathway (PPP) and glycolysis pathways resulted in better ethanol production in S. stipitis and S. passalidarum when compared to S. arborariae.
Keywords: Cofactor balance; Ethanol; MFA; Metabolome; Metabolomics; Xylose metabolism.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures




References
-
- du Preez JC, van Driessel B, Prior BA. D-xylose fermentation by Candida shehatae and Pichia stipitis at low dissolved oxygen levels in fed-batch cultures. Biotechnol Lett. 1989;11:131–136. doi: 10.1007/BF01192189. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

