An RNA Aptamer-Based Biomarker Platform Demonstrates High Soluble CD25 Occupancy by IL2 in the Serum of Follicular Lymphoma Patients
- PMID: 31383650
- PMCID: PMC6726511
- DOI: 10.1158/2326-6066.CIR-18-0821
An RNA Aptamer-Based Biomarker Platform Demonstrates High Soluble CD25 Occupancy by IL2 in the Serum of Follicular Lymphoma Patients
Abstract
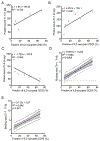
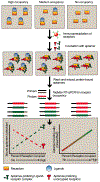
Ligand-receptor complexes play a central role in mediating a range of processes in immunology and cancer biology. The ability to directly quantify the fraction of receptors occupied by a ligand in a given biospecimen, as opposed to assessing the concentration of ligand and receptor separately, could provide an additional and valuable clinical and research tool for assessing whether receptors are occupied by a ligand. To address this need, a biomarker platform was developed to quantify the fraction of receptors occupied by a ligand using pairs of RNA aptamers, where one aptamer binds preferentially to the unoccupied receptor and the other to the ligand-receptor complex. Bound aptamer was quantified using RT-qPCR colorimetric probes specific for each aptamer. The binding ratio of aptamer correlated with the fraction of receptors occupied by a ligand. This assay, termed as LIRECAP (LIgand-REceptor Complex-binding APtamer) assay, was used to determine the fraction of soluble CD25 occupied by IL2 in the serum from subjects with B-cell lymphoma. No correlation was found between the type of lymphoma and total soluble CD25 or IL2 independently. In contrast, the fraction of soluble CD25 occupied by IL2 was significantly higher in follicular lymphoma patient serum compared with diffuse large B-cell lymphoma patient serum. We conclude that this technology has the potential to serve as a high-throughput biomarker platform to quantify the fraction of receptors occupied by a ligand.
©2019 American Association for Cancer Research.
Conflict of interest statement
Competing financial interests
The authors declare no competing financial interests.
Figures

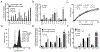


Similar articles
-
Quantification of Receptor Occupancy by Ligand-An Understudied Class of Potential Biomarkers.Cancers (Basel). 2020 Oct 13;12(10):2956. doi: 10.3390/cancers12102956. Cancers (Basel). 2020. PMID: 33066142 Free PMC article.
-
Design, isolation and evaluation of the binding efficiency of a DNA aptamer against interleukin 2 receptor alpha, in vitro.Int Immunopharmacol. 2017 Dec;53:96-104. doi: 10.1016/j.intimp.2017.10.011. Epub 2017 Oct 18. Int Immunopharmacol. 2017. PMID: 29055191
-
Evolution of Cell-Type-Specific RNA Aptamers Via Live Cell-Based SELEX.Methods Mol Biol. 2016;1421:191-214. doi: 10.1007/978-1-4939-3591-8_16. Methods Mol Biol. 2016. PMID: 26965267
-
Recent developments in cell-SELEX technology for aptamer selection.Biochim Biophys Acta Gen Subj. 2018 Oct;1862(10):2323-2329. doi: 10.1016/j.bbagen.2018.07.029. Epub 2018 Jul 27. Biochim Biophys Acta Gen Subj. 2018. PMID: 30059712 Review.
-
Cancer protein biomarker discovery based on nucleic acid aptamers.Int J Biol Macromol. 2019 Jul 1;132:190-202. doi: 10.1016/j.ijbiomac.2019.03.165. Epub 2019 Mar 26. Int J Biol Macromol. 2019. PMID: 30926499 Review.
Cited by
-
Phenotype, Function, and Clinical Significance of CD26+ and CD161+Tregs in Splenic Marginal Zone Lymphoma.Clin Cancer Res. 2022 Oct 3;28(19):4322-4335. doi: 10.1158/1078-0432.CCR-22-0977. Clin Cancer Res. 2022. PMID: 35686915 Free PMC article.
-
Selection of a novel cell-internalizing RNA aptamer specific for CD22 antigen in B cell acute lymphoblastic leukemia.Mol Ther Nucleic Acids. 2023 Jul 28;33:698-712. doi: 10.1016/j.omtn.2023.07.028. eCollection 2023 Sep 12. Mol Ther Nucleic Acids. 2023. PMID: 37662970 Free PMC article.
-
Quantification of Receptor Occupancy by Ligand-An Understudied Class of Potential Biomarkers.Cancers (Basel). 2020 Oct 13;12(10):2956. doi: 10.3390/cancers12102956. Cancers (Basel). 2020. PMID: 33066142 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

