Production of Norspermidine Contributes to Aminoglycoside Resistance in pmrAB Mutants of Pseudomonas aeruginosa
- PMID: 31383668
- PMCID: PMC6761568
- DOI: 10.1128/AAC.01044-19
Production of Norspermidine Contributes to Aminoglycoside Resistance in pmrAB Mutants of Pseudomonas aeruginosa
Abstract
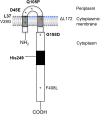
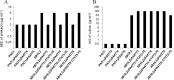
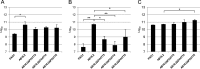
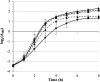
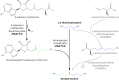
Emergence of resistance to polymyxins in Pseudomonas aeruginosa is mainly due to mutations in two-component systems that promote the addition of 4-amino-4-deoxy-l-arabinose to the lipopolysaccharide (LPS) through upregulation of operon arnBCADTEF-ugd (arn) expression. Here, we demonstrate that mutations occurring in different domains of histidine kinase PmrB or in response regulator PmrA result in coresistance to aminoglycosides and colistin. All seventeen clinical strains tested exhibiting such a cross-resistance phenotype were found to be pmrAB mutants. As shown by gene deletion experiments, the decreased susceptibility of the mutants to aminoglycosides was independent from operon arn but required the efflux system MexXY-OprM and the products of three genes, PA4773-PA4774-PA4775, that are cotranscribed and activated with genes pmrAB Gene PA4773 (annotated as speD2 in the PAO1 genome) and PA4774 (speE2) are predicted to encode enzymes involved in biosynthesis of polyamines. Comparative analysis of cell surface extracts of an in vitro selected pmrAB mutant, called AB16.2, and derivatives lacking PA4773, PA4774, and PA4775 revealed that these genes were needed for norspermidine production via a pathway that likely uses 1,3-diaminopropane, a precursor of polyamines. Altogether, our results suggest that norspermidine decreases the self-promoted uptake pathway of aminoglycosides across the outer membrane and, thereby, potentiates the activity of efflux pump MexXY-OprM.
Keywords: PmrAB; Pseudomonas aeruginosa; aminoglycosides; colistin; norspermidine; polyamines.
Copyright © 2019 American Society for Microbiology.
Figures
References
-
- Del Barrio-Tofino E, Lopez-Causapé C, Cabot G, Rivera A, Benito N, Segura C, Montero MM, Sorli L, Tubau F, Gomez-Zorrilla S, Tormo N, Dura-Navarro R, Viedma E, Resino-Foz E, Fernandez-Martinez M, Gonzalez-Rico C, Alejo-Cancho I, Martinez JA, Labayru-Echverria C, Duenas C, Ayestaran I, Zamorano L, Martinez-Martinez L, Horcajada JP, Oliver A. 2017. Genomics and susceptibility profiles of extensively drug-resistant Pseudomonas aeruginosa isolates from Spain. Antimicrob Agents Chemother 61:e01589-17. doi: 10.1128/AAC.01589-17. - DOI - PMC - PubMed
-
- Sader HS, Huband MD, Castanheira M, Flamm RK. 2017. Pseudomonas aeruginosa antimicrobial susceptibility results from four years (2012 to 2015) of the international network for optimal resistance monitoring program in the United States. Antimicrob Agents Chemother 61:e02252-16. doi: 10.1128/AAC.02252-16. - DOI - PMC - PubMed
-
- Nummila K, Kilpelainen I, Zahringer U, Vaara M, Helander IM. 1995. Lipopolysaccharides of polymyxin B-resistant mutants of Escherichia coli are extensively substituted by 2-aminoethyl pyrophosphate and contain aminoarabinose in lipid A. Mol Microbiol 16:271–278. doi: 10.1111/j.1365-2958.1995.tb02299.x. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

