Quantifying the impact of public omics data
- PMID: 31383865
- PMCID: PMC6683138
- DOI: 10.1038/s41467-019-11461-w
Quantifying the impact of public omics data
Abstract
The amount of omics data in the public domain is increasing every year. Modern science has become a data-intensive discipline. Innovative solutions for data management, data sharing, and for discovering novel datasets are therefore increasingly required. In 2016, we released the first version of the Omics Discovery Index (OmicsDI) as a light-weight system to aggregate datasets across multiple public omics data resources. OmicsDI aggregates genomics, transcriptomics, proteomics, metabolomics and multiomics datasets, as well as computational models of biological processes. Here, we propose a set of novel metrics to quantify the attention and impact of biomedical datasets. A complete framework (now integrated into OmicsDI) has been implemented in order to provide and evaluate those metrics. Finally, we propose a set of recommendations for authors, journals and data resources to promote an optimal quantification of the impact of datasets.
Conflict of interest statement
The authors declare no competing interests.
Figures


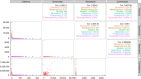
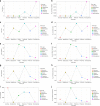
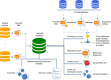

References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

