Structure functional insights into calcium binding during the activation of coagulation factor XIII A
- PMID: 31383913
- PMCID: PMC6683118
- DOI: 10.1038/s41598-019-47815-z
Structure functional insights into calcium binding during the activation of coagulation factor XIII A
Abstract
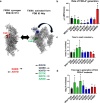
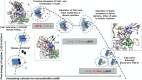
The dimeric FXIII-A2, a pro-transglutaminase is the catalytic part of the heterotetrameric coagulation FXIII-A2B2 complex that upon activation by calcium binding/thrombin cleavage covalently cross-links preformed fibrin clots protecting them from premature fibrinolysis. Our study characterizes the recently disclosed three calcium binding sites of FXIII-A concerning evolution, mutual crosstalk, thermodynamic activation profile, substrate binding, and interaction with other similarly charged ions. We demonstrate unique structural aspects within FXIII-A calcium binding sites that give rise to functional differences making FXIII unique from other transglutaminases. The first calcium binding site showed an antagonistic relationship towards the other two. The thermodynamic profile of calcium/thrombin-induced FXIII-A activation explains the role of bulk solvent in transitioning its zymogenic dimeric form to an activated monomeric form. We also explain the indirect effect of solvent ion concentration on FXIII-A activation. Our study suggests FXIII-A calcium binding sites could be putative pharmacologically targetable regions.
Conflict of interest statement
The authors declare no competing interests.
Figures






References
-
- Lorand L, Losowsky MS, Miloszewski KJ. Human factor XIII: fibrin-stabilizing factor. Progress in hemostasis and thrombosis. 1980;5:245–290. - PubMed
-
- Schwartz ML, Pizzo SV, Hill RL, McKee PA. Human Factor XIII from plasma and platelets. Molecular weights, subunit structures, proteolytic activation, and cross-linking of fibrinogen and fibrin. The Journal of biological chemistry. 1973;248:1395–1407. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

