Selection for antimicrobial resistance is reduced when embedded in a natural microbial community
- PMID: 31384011
- PMCID: PMC6864104
- DOI: 10.1038/s41396-019-0483-z
Selection for antimicrobial resistance is reduced when embedded in a natural microbial community
Abstract
Antibiotic resistance has emerged as one of the most pressing, global threats to public health. In single-species experiments selection for antibiotic resistance occurs at very low antibiotic concentrations. However, it is unclear how far these findings can be extrapolated to natural environments, where species are embedded within complex communities. We competed isogenic strains of Escherichia coli, differing exclusively in a single chromosomal resistance determinant, in the presence and absence of a pig faecal microbial community across a gradient of antibiotic concentration for two relevant antibiotics: gentamicin and kanamycin. We show that the minimal selective concentration was increased by more than one order of magnitude for both antibiotics when embedded in the community. We identified two general mechanisms were responsible for the increase in minimal selective concentration: an increase in the cost of resistance and a protective effect of the community for the susceptible phenotype. These findings have implications for our understanding of the evolution and selection of antibiotic resistance, and can inform future risk assessment efforts on antibiotic concentrations.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures



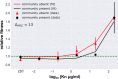
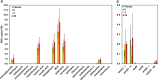
Comment in
-
Microbial Ecology: Complex Bacterial Communities Reduce Selection for Antibiotic Resistance.Curr Biol. 2019 Nov 4;29(21):R1143-R1145. doi: 10.1016/j.cub.2019.09.017. Curr Biol. 2019. PMID: 31689403
References
-
- WHO. Antimicrobial Resistance Global Report on Surveillance. 2014. https://www.who.int/drugresistance/documents/surveillancereport/en/.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

