Glycerol phosphate shuttle enzyme GPD2 regulates macrophage inflammatory responses
- PMID: 31384058
- PMCID: PMC6707851
- DOI: 10.1038/s41590-019-0453-7
Glycerol phosphate shuttle enzyme GPD2 regulates macrophage inflammatory responses
Erratum in
-
Author Correction: Glycerol phosphate shuttle enzyme GPD2 regulates macrophage inflammatory responses.Nat Immunol. 2019 Nov;20(11):1555. doi: 10.1038/s41590-019-0517-8. Nat Immunol. 2019. PMID: 31551573
Abstract
Macrophages are activated during microbial infection to coordinate inflammatory responses and host defense. Here we find that in macrophages activated by bacterial lipopolysaccharide (LPS), mitochondrial glycerol 3-phosphate dehydrogenase (GPD2) regulates glucose oxidation to drive inflammatory responses. GPD2, a component of the glycerol phosphate shuttle, boosts glucose oxidation to fuel the production of acetyl coenzyme A, acetylation of histones and induction of genes encoding inflammatory mediators. While acute exposure to LPS drives macrophage activation, prolonged exposure to LPS triggers tolerance to LPS, where macrophages induce immunosuppression to limit the detrimental effects of sustained inflammation. The shift in the inflammatory response is modulated by GPD2, which coordinates a shutdown of oxidative metabolism; this limits the availability of acetyl coenzyme A for histone acetylation at genes encoding inflammatory mediators and thus contributes to the suppression of inflammatory responses. Therefore, GPD2 and the glycerol phosphate shuttle integrate the extent of microbial stimulation with glucose oxidation to balance the beneficial and detrimental effects of the inflammatory response.
Conflict of interest statement
Competing Interests
The authors declare no competing interests.
Figures
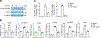
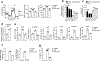
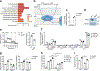
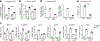
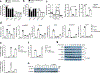

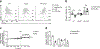
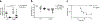
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

