Cytomegalovirus distribution and evolution in hominines
- PMID: 31384482
- PMCID: PMC6671425
- DOI: 10.1093/ve/vez015
Cytomegalovirus distribution and evolution in hominines
Abstract
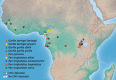
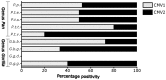
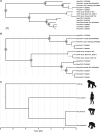
Herpesviruses are thought to have evolved in very close association with their hosts. This is notably the case for cytomegaloviruses (CMVs; genus Cytomegalovirus) infecting primates, which exhibit a strong signal of co-divergence with their hosts. Some herpesviruses are however known to have crossed species barriers. Based on a limited sampling of CMV diversity in the hominine (African great ape and human) lineage, we hypothesized that chimpanzees and gorillas might have mutually exchanged CMVs in the past. Here, we performed a comprehensive molecular screening of all 9 African great ape species/subspecies, using 675 fecal samples collected from wild animals. We identified CMVs in eight species/subspecies, notably generating the first CMV sequences from bonobos. We used this extended dataset to test competing hypotheses with various degrees of co-divergence/number of host switches while simultaneously estimating the dates of these events in a Bayesian framework. The model best supported by the data involved the transmission of a gorilla CMV to the panine (chimpanzee and bonobo) lineage and the transmission of a panine CMV to the gorilla lineage prior to the divergence of chimpanzees and bonobos, more than 800,000 years ago. Panine CMVs then co-diverged with their hosts. These results add to a growing body of evidence suggesting that viruses with a double-stranded DNA genome (including other herpesviruses, adenoviruses, and papillomaviruses) often jumped between hominine lineages over the last few million years.
Keywords: codivergence; cytomegalovirus; dsDNA virus; hominine; host switch.
Figures


References
-
- Altschul S. F. et al. (1990) ‘Basic Local Alignment Search Tool’, Journal of Molecular Biology, 215: 403–10. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

