Integrated genetic and epigenetic analysis revealed heterogeneity of acute lymphoblastic leukemia in Down syndrome
- PMID: 31385395
- PMCID: PMC6778645
- DOI: 10.1111/cas.14160
Integrated genetic and epigenetic analysis revealed heterogeneity of acute lymphoblastic leukemia in Down syndrome
Abstract
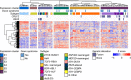
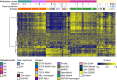
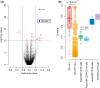
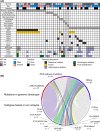
Children with Down syndrome (DS) are at a 20-fold increased risk for acute lymphoblastic leukemia (ALL). Compared to children with ALL and no DS (non-DS-ALL), those with DS and ALL (DS-ALL) harbor uncommon genetic alterations, suggesting DS-ALL could have distinct biological features. Recent studies have implicated several genes on chromosome 21 in DS-ALL, but the precise mechanisms predisposing children with DS to ALL remain unknown. Our integrated genetic/epigenetic analysis revealed that DS-ALL was highly heterogeneous with many subtypes. Although each subtype had genetic/epigenetic profiles similar to those found in non-DS-ALL, the subtype distribution differed significantly between groups. The Philadelphia chromosome-like subtype, a high-risk B-cell lineage variant relatively rare among the entire pediatric ALL population, was the most common form in DS-ALL. Hypermethylation of RUNX1 on chromosome 21 was also found in DS-ALL, but not non-DS-ALL. RUNX1 is essential for differentiation of blood cells, especially B cells; thus, hypermethylation of the RUNX1 promoter in B-cell precursors might be associated with increased incidence of B-cell precursor ALL in DS patients.
Keywords: Down syndrome; acute lymphoblastic leukemia; children; epigenetic analysis; genetic analysis.
© 2019 The Authors. Cancer Science published by John Wiley & Sons Australia, Ltd on behalf of Japanese Cancer Association.
Figures

References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

