The central role of the tail in switching off 10S myosin II activity
- PMID: 31387899
- PMCID: PMC6719407
- DOI: 10.1085/jgp.201912431
The central role of the tail in switching off 10S myosin II activity
Abstract
Myosin II is a motor protein with two heads and an extended tail that plays an essential role in cell motility. Its active form is a polymer (myosin filament) that pulls on actin to generate motion. Its inactive form is a monomer with a compact structure (10S sedimentation coefficient), in which the tail is folded and the two heads interact with each other, inhibiting activity. This conformation is thought to function in cells as an energy-conserving form of the molecule suitable for storage as well as transport to sites of filament assembly. The mechanism of inhibition of the compact molecule is not fully understood. We have performed a 3-D reconstruction of negatively stained 10S myosin from smooth muscle in the inhibited state using single-particle analysis. The reconstruction reveals multiple interactions between the tail and the two heads that appear to trap ATP hydrolysis products, block actin binding, hinder head phosphorylation, and prevent filament formation. Blocking these essential features of myosin function could explain the high degree of inhibition of the folded form of myosin thought to underlie its energy-conserving function in cells. The reconstruction also suggests a mechanism for unfolding when myosin is activated by phosphorylation.
© 2019 Yang et al.
Figures
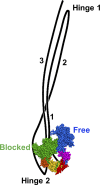


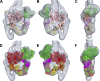
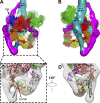
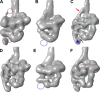
Comment in
-
How myosin II achieves total shutdown.J Gen Physiol. 2019 Sep 2;151(9):1061. doi: 10.1085/jgp.201912458. Epub 2019 Aug 8. J Gen Physiol. 2019. PMID: 31395613 Free PMC article.
References
-
- Alamo L., Qi D., Wriggers W., Pinto A., Zhu J., Bilbao A., Gillilan R.E., Hu S., and Padrón R.. 2016. Conserved Intramolecular Interactions Maintain Myosin Interacting-Heads Motifs Explaining Tarantula Muscle Super-Relaxed State Structural Basis. J. Mol. Biol. 428:1142–1164. 10.1016/j.jmb.2016.01.027 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources

