Sulfur Assimilation Alters Flagellar Function and Modulates the Gene Expression Landscape of Serratia marcescens
- PMID: 31387930
- PMCID: PMC6687942
- DOI: 10.1128/mSystems.00285-19
Sulfur Assimilation Alters Flagellar Function and Modulates the Gene Expression Landscape of Serratia marcescens
Abstract
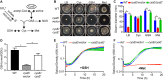
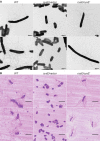
Sulfur is an essential nutrient that contributes to cellular redox homeostasis, transcriptional regulation, and translation initiation when incorporated into different biomolecules. Transport and reduction of extracellular sulfate followed by cysteine biosynthesis is a major pathway of bacterial sulfur assimilation. For the opportunistic pathogen Serratia marcescens, function of the cysteine biosynthesis pathway is required for extracellular phospholipase activity and flagellum-mediated surface motility, but little else is known about the influence of sulfur assimilation on the physiology of this organism. In this work, it was determined that an S. marcescens cysteine auxotroph fails to differentiate into hyperflagellated and elongated swarmer cells and that cysteine, but not other organic sulfur molecules, restores swarming motility to these bacteria. The S. marcescens cysteine auxotroph further exhibits reduced transcription of phospholipase, hemolysin, and flagellin genes, each of which is subject to transcriptional control by the flagellar regulatory system. Based on these data and the central role of cysteine in sulfur assimilation, it was reasoned that environmental sulfur availability may contribute to the regulation of these functions in S. marcescens Indeed, bacteria that are starved for sulfate exhibit substantially reduced transcription of the genes for hemolysin, phospholipase, and the FlhD flagellar master regulator. A global transcriptomic analysis further defined a large set of S. marcescens genes that are responsive to extracellular sulfate availability, including genes that encode membrane transport, nutrient utilization, and metabolism functions. Finally, sulfate availability was demonstrated to alter S. marcescens cytolytic activity, suggesting that sulfate assimilation may impact the virulence of this organism.IMPORTANCE Serratia marcescens is a versatile bacterial species that inhabits diverse environmental niches and is capable of pathogenic interactions with host organisms ranging from insects to humans. This report demonstrates for the first time the extensive impacts that environmental sulfate availability and cysteine biosynthesis have on the transcriptome of S. marcescens The finding that greater than 1,000 S. marcescens genes are differentially expressed depending on sulfate availability suggests that sulfur abundance is a crucial factor that controls the physiology of this organism. Furthermore, the high relative expression levels for the putative virulence factors flagella, phospholipase, and hemolysin in the presence of sulfate suggests that a sulfur-rich host environment could contribute to the transcription of these genes during infection.
Keywords: Serratia; cysteine; flagella; hemolysin; phospholipase; sulfur.
Copyright © 2019 Anderson et al.
Figures





References
-
- Kredich NM, Tomkins GM. 1966. The enzymic synthesis of l-cysteine in Escherichia coli and Salmonella Typhimurium. J Biol Chem 241:4955–4965. - PubMed
-
- Hryniewicz MM, Kredich NM. 1991. The cysP promoter of Salmonella Typhimurium: characterization of two binding sites for CysB protein, studies of in vivo transcription initiation, and demonstration of the anti-inducer effects of thiosulfate. J Bacteriol 173:5876–5886. doi:10.1128/jb.173.18.5876-5886.1991. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

