Longitudinal Comparison of Bacterial Diversity and Antibiotic Resistance Genes in New York City Sewage
- PMID: 31387933
- PMCID: PMC6687945
- DOI: 10.1128/mSystems.00327-19
Longitudinal Comparison of Bacterial Diversity and Antibiotic Resistance Genes in New York City Sewage
Abstract
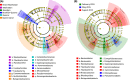
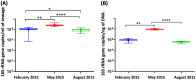
Bacterial resistance to antibiotics is a pressing health issue around the world, not only in health care settings but also in the community and environment, particularly in crowded urban populations. The aim of our work was to characterize the microbial populations in sewage and the spread of antibiotic resistance within New York City (NYC). Here, we investigated the structure of the microbiome and the prevalence of antibiotic resistance genes in raw sewage samples collected from the fourteen NYC Department of Environmental Protection wastewater treatment plants, distributed across the five NYC boroughs. Sewage, a direct output of anthropogenic activity and a reservoir of microbes, provides an ecological niche to examine the spread of antibiotic resistance. Taxonomic diversity analysis revealed a largely similar and stable bacterial population structure across all the samples, which was found to be similar over three time points in an annual cycle, as well as in the five NYC boroughs. All samples were positive for the presence of the seven antibiotic resistance genes tested, based on real-time quantitative PCR assays, with higher levels observed for tetracycline resistance genes at all time points. For five of the seven genes, abundances were significantly higher in May than in February and August. This study provides characteristics of the NYC sewage resistome in the context of the overall bacterial populations.IMPORTANCE Urban sewage or wastewater is a diverse source of bacterial growth, as well as a hot spot for the development of environmental antibiotic resistance, which can in turn influence the health of the residents of the city. As part of a larger study to characterize the urban New York City microbial metagenome, we collected raw sewage samples representing three seasonal time points spanning the five boroughs of NYC and went on to characterize the microbiome and the presence of a range of antibiotic resistance genes. Through this study, we have established a baseline microbial population and antibiotic resistance abundance in NYC sewage which can prove to be very useful in studying the load of antibiotic usage, as well as for developing effective measures in antibiotic stewardship.
Keywords: New York City; antibiotic resistance; microbiome; sewage.
Copyright © 2019 Joseph et al.
Figures





References
-
- Laxminarayan R, Duse A, Wattal C, Zaidi AKM, Wertheim HFL, Sumpradit N, Vlieghe E, Hara GL, Gould IM, Goossens H, Greko C, So AD, Bigdeli M, Tomson G, Woodhouse W, Ombaka E, Peralta AQ, Qamar FN, Mir F, Kariuki S, Bhutta ZA, Coates A, Bergstrom R, Wright GD, Brown ED, Cars O. 2013. Antibiotic resistance: the need for global solutions. Lancet Infect Dis 13:1057–1098. doi: 10.1016/S1473-3099(13)70318-9. - DOI - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

