BIK ubiquitination by the E3 ligase Cul5-ASB11 determines cell fate during cellular stress
- PMID: 31387940
- PMCID: PMC6719446
- DOI: 10.1083/jcb.201901156
BIK ubiquitination by the E3 ligase Cul5-ASB11 determines cell fate during cellular stress
Abstract
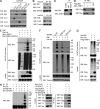
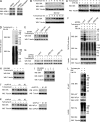
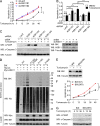
The BH3-only pro-apoptotic protein BIK is regulated by the ubiquitin-proteasome system. However, the mechanism of this regulation and its physiological functions remain elusive. Here, we identify Cul5-ASB11 as the E3 ligase targeting BIK for ubiquitination and degradation. ER stress leads to the activation of ASB11 by XBP1s during the adaptive phase of the unfolded protein response, which stimulates BIK ubiquitination, interaction with p97/VCP, and proteolysis. This mechanism of BIK degradation contributes to ER stress adaptation by promoting cell survival. Conversely, genotoxic agents down-regulate this IRE1α-XBP1s-ASB11 axis and stabilize BIK, which contributes in part to the apoptotic response to DNA damage. We show that blockade of this BIK degradation pathway by an IRE1α inhibitor can stabilize a BIK active mutant and increase its anti-tumor activity. Our study reveals that different cellular stresses regulate BIK ubiquitination by ASB11 in opposing directions, which determines whether or not cells survive, and that blocking BIK degradation has the potential to be used as an anti-cancer strategy.
© 2019 Chen et al.
Figures





References
-
- Andresen C.A., Smedegaard S., Sylvestersen K.B., Svensson C., Iglesias-Gato D., Cazzamali G., Nielsen T.K., Nielsen M.L., and Flores-Morales A.. 2014. Protein interaction screening for the ankyrin repeats and suppressor of cytokine signaling (SOCS) box (ASB) family identify Asb11 as a novel endoplasmic reticulum resident ubiquitin ligase. J. Biol. Chem. 289:2043–2054. 10.1074/jbc.M113.534602 - DOI - PMC - PubMed
-
- Boyd J.M., Gallo G.J., Elangovan B., Houghton A.B., Malstrom S., Avery B.J., Ebb R.G., Subramanian T., Chittenden T., Lutz R.J., et al. 1995. Bik, a novel death-inducing protein shares a distinct sequence motif with Bcl-2 family proteins and interacts with viral and cellular survival-promoting proteins. Oncogene. 11:1921–1928. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

