Mutational processes contributing to the development of multiple myeloma
- PMID: 31387987
- PMCID: PMC6684612
- DOI: 10.1038/s41408-019-0221-9
Mutational processes contributing to the development of multiple myeloma
Abstract
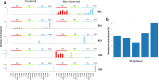
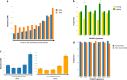
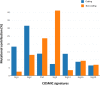
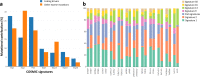
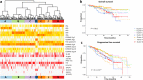
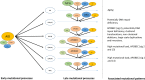
To gain insight into multiple myeloma (MM) tumorigenesis, we analyzed the mutational signatures in 874 whole-exome and 850 whole-genome data from the CoMMpass Study. We identified that coding and non-coding regions are differentially dominated by distinct single-nucleotide variant (SNV) mutational signatures, as well as five de novo structural rearrangement signatures. Mutational signatures reflective of different principle mutational processes-aging, defective DNA repair, and apolipoprotein B editing complex (APOBEC)/activation-induced deaminase activity-characterize MM. These mutational signatures show evidence of subgroup specificity-APOBEC-attributed signatures associated with MAF translocation t(14;16) and t(14;20) MM; potentially DNA repair deficiency with t(11;14) and t(4;14); and aging with hyperdiploidy. Mutational signatures beyond that associated with APOBEC are independent of established prognostic markers and appear to have relevance to predicting high-risk MM.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

