Incomplete influenza A virus genomes occur frequently but are readily complemented during localized viral spread
- PMID: 31387995
- PMCID: PMC6684657
- DOI: 10.1038/s41467-019-11428-x
Incomplete influenza A virus genomes occur frequently but are readily complemented during localized viral spread
Abstract
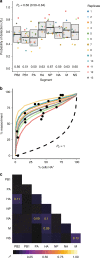
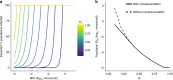
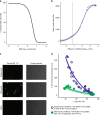
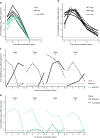
Segmentation of viral genomes into multiple RNAs creates the potential for replication of incomplete viral genomes (IVGs). Here we use a single-cell approach to quantify influenza A virus IVGs and examine their fitness implications. We find that each segment of influenza A/Panama/2007/99 (H3N2) virus has a 58% probability of being replicated in a cell infected with a single virion. Theoretical methods predict that IVGs carry high costs in a well-mixed system, as 3.6 virions are required for replication of a full genome. Spatial structure is predicted to mitigate these costs, however, and experimental manipulations of spatial structure indicate that local spread facilitates complementation. A virus entirely dependent on co-infection was used to assess relevance of IVGs in vivo. This virus grows robustly in guinea pigs, but is less infectious and does not transmit. Thus, co-infection allows IVGs to contribute to within-host spread, but complete genomes may be critical for transmission.
Conflict of interest statement
The authors declare no competing interests.
Figures




Similar articles
-
A Novel Type of Influenza A Virus-Derived Defective Interfering Particle with Nucleotide Substitutions in Its Genome.J Virol. 2019 Feb 5;93(4):e01786-18. doi: 10.1128/JVI.01786-18. Print 2019 Feb 15. J Virol. 2019. PMID: 30463972 Free PMC article.
-
Inhibition of Ongoing Influenza A Virus Replication Reveals Different Mechanisms of RIG-I Activation.J Virol. 2019 Mar 5;93(6):e02066-18. doi: 10.1128/JVI.02066-18. Print 2019 Mar 15. J Virol. 2019. PMID: 30602605 Free PMC article.
-
Antiviral resistance due to deletion in the neuraminidase gene and defective interfering-like viral polymerase basic 2 RNA of influenza A virus subtype H3N2.J Clin Virol. 2018 May;102:1-6. doi: 10.1016/j.jcv.2018.02.005. Epub 2018 Feb 11. J Clin Virol. 2018. PMID: 29448067
-
Implications of segment mismatch for influenza A virus evolution.J Gen Virol. 2018 Jan;99(1):3-16. doi: 10.1099/jgv.0.000989. Epub 2017 Dec 15. J Gen Virol. 2018. PMID: 29244017 Free PMC article. Review.
-
Moving On Out: Transport and Packaging of Influenza Viral RNA into Virions.Annu Rev Virol. 2016 Sep 29;3(1):411-427. doi: 10.1146/annurev-virology-110615-042345. Annu Rev Virol. 2016. PMID: 27741407 Review.
Cited by
-
High-throughput droplet-based analysis of influenza A virus genetic reassortment by single-virus RNA sequencing.Proc Natl Acad Sci U S A. 2023 Feb 7;120(6):e2211098120. doi: 10.1073/pnas.2211098120. Epub 2023 Feb 2. Proc Natl Acad Sci U S A. 2023. PMID: 36730204 Free PMC article.
-
Influenza A virus undergoes compartmentalized replication in vivo dominated by stochastic bottlenecks.Nat Commun. 2022 Jun 14;13(1):3416. doi: 10.1038/s41467-022-31147-0. Nat Commun. 2022. PMID: 35701424 Free PMC article.
-
Modeling recapitulates the heterogeneous outcomes of SARS-CoV-2 infection and quantifies the differences in the innate immune and CD8 T-cell responses between patients experiencing mild and severe symptoms.PLoS Pathog. 2022 Jun 27;18(6):e1010630. doi: 10.1371/journal.ppat.1010630. eCollection 2022 Jun. PLoS Pathog. 2022. PMID: 35759522 Free PMC article.
-
Cooperative Virus-Virus Interactions: An Evolutionary Perspective.Biodes Res. 2022 Aug 9;2022:9819272. doi: 10.34133/2022/9819272. eCollection 2022. Biodes Res. 2022. PMID: 37850129 Free PMC article. Review.
-
Rab11a mediates cell-cell spread and reassortment of influenza A virus genomes via tunneling nanotubes.PLoS Pathog. 2021 Sep 2;17(9):e1009321. doi: 10.1371/journal.ppat.1009321. eCollection 2021 Sep. PLoS Pathog. 2021. PMID: 34473799 Free PMC article.
References
-
- Chao L, Tran TR, Matthews C. Muller ratchet and the advantage of sex in the Rna Virus-Phi-6. Evol.; Int. J. Org. Evol. 1992;46:289–299. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

