Incomplete influenza A virus genomes occur frequently but are readily complemented during localized viral spread
- PMID: 31387995
- PMCID: PMC6684657
- DOI: 10.1038/s41467-019-11428-x
Incomplete influenza A virus genomes occur frequently but are readily complemented during localized viral spread
Abstract
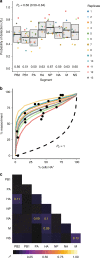
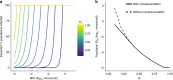
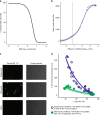
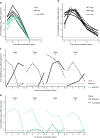
Segmentation of viral genomes into multiple RNAs creates the potential for replication of incomplete viral genomes (IVGs). Here we use a single-cell approach to quantify influenza A virus IVGs and examine their fitness implications. We find that each segment of influenza A/Panama/2007/99 (H3N2) virus has a 58% probability of being replicated in a cell infected with a single virion. Theoretical methods predict that IVGs carry high costs in a well-mixed system, as 3.6 virions are required for replication of a full genome. Spatial structure is predicted to mitigate these costs, however, and experimental manipulations of spatial structure indicate that local spread facilitates complementation. A virus entirely dependent on co-infection was used to assess relevance of IVGs in vivo. This virus grows robustly in guinea pigs, but is less infectious and does not transmit. Thus, co-infection allows IVGs to contribute to within-host spread, but complete genomes may be critical for transmission.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- Chao L, Tran TR, Matthews C. Muller ratchet and the advantage of sex in the Rna Virus-Phi-6. Evol.; Int. J. Org. Evol. 1992;46:289–299. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

