Nucleosomes Regulate Base Excision Repair in Chromatin
- PMID: 31388331
- PMCID: PMC6684245
- DOI: 10.1016/j.mrrev.2017.10.002
Nucleosomes Regulate Base Excision Repair in Chromatin
Abstract
Chromatin is a significant barrier to many DNA damage response (DDR) factors, such as DNA repair enzymes, that process DNA lesions to reduce mutations and prevent cell death; yet, paradoxically, chromatin also has a critical role in many signaling pathways that regulate the DDR. The primary level of DNA packaging in chromatin is the nucleosome core particle (NCP), consisting of DNA wrapped around an octamer of the core histones H2A, H2B, H3 and H4. Here, we review recent studies characterizing how the packaging of DNA into nucleosomes modulates the activity of the base excision repair (BER) pathway and dictates BER subpathway choice. We also review new evidence indicating that the histone amino-terminal tails coordinately regulate multiple DDR pathways during the repair of alkylation damage in the budding yeast Saccharomyces cerevisiae.
Figures

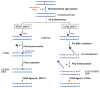
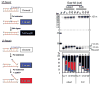


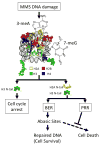
References
-
- Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenberger T. DNA repair and mutagenesis. 2. ASM Press; Washington, D.C: 2006.
-
- Bernstein C, Prassad AR, Nfonsam V, Bernstein H. DNA Damage, DNA Repair and Cancer. In: Chen CC, editor. New Research Directions. Intech, open access; 2013. pp. 413–465.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
