Constructing a Reference Genome in a Single Lab: The Possibility to Use Oxford Nanopore Technology
- PMID: 31390788
- PMCID: PMC6724115
- DOI: 10.3390/plants8080270
Constructing a Reference Genome in a Single Lab: The Possibility to Use Oxford Nanopore Technology
Abstract
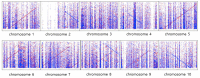
The whole genome sequencing (WGS) has become a crucial tool in understanding genome structure and genetic variation. The MinION sequencing of Oxford Nanopore Technologies (ONT) is an excellent approach for performing WGS and it has advantages in comparison with other Next-Generation Sequencing (NGS): It is relatively inexpensive, portable, has simple library preparation, can be monitored in real-time, and has no theoretical limits on reading length. Sorghum bicolor (L.) Moench is diploid (2n = 2x = 20) with a genome size of about 730 Mb, and its genome sequence information is released in the Phytozome database. Therefore, sorghum can be used as a good reference. However, plant species have complex and large genomes when compared to animals or microorganisms. As a result, complete genome sequencing is difficult for plant species. MinION sequencing that produces long-reads can be an excellent tool for overcoming the weak assembly of short-reads generated from NGS by minimizing the generation of gaps or covering the repetitive sequence that appears on the plant genome. Here, we conducted the genome sequencing for S. bicolor cv. BTx623 while using the MinION platform and obtained 895,678 reads and 17.9 gigabytes (Gb) (ca. 25× coverage of reference) from long-read sequence data. A total of 6124 contigs (covering 45.9%) were generated from Canu, and a total of 2661 contigs (covering 50%) were generated from Minimap and Miniasm with a Racon through a de novo assembly using two different tools and mapped assembled contigs against the sorghum reference genome. Our results provide an optimal series of long-read sequencing analysis for plant species while using the MinION platform and a clue to determine the total sequencing scale for optimal coverage that is based on various genome sizes.
Keywords: Canu; Keywords: sorghum; MinION; Miniasm; long-read sequencing.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

References
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

