miR-448 targets IDO1 and regulates CD8+ T cell response in human colon cancer
- PMID: 31391111
- PMCID: PMC6686234
- DOI: 10.1186/s40425-019-0691-0
miR-448 targets IDO1 and regulates CD8+ T cell response in human colon cancer
Abstract
Background: Indoleamine 2,3-dioxygenase 1 (IDO1) is a critical regulator of T cell function, contributing to immune tolerance. Upregulation of IDO1 has been found in many cancer types; however, the regulatory mechanisms and clinical significance of IDO1 in colon cancer are still unclear. Here, we investigated the role of dysregulated microRNA (miRNA) targeting IDO1 in the colon cancer microenvironment.
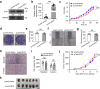
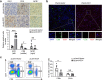
Methods: We elucidated IDO1 function by performing cell-based assays and establishing transplanted tumor models in BALB/c mice and BALB/c nude mice. We evaluated IDO1 protein expression by immunohistochemistry (IHC) in a tissue microarray (TMA) and analyzed IDO1 mRNA expression with The Cancer Genome Atlas (TCGA). We screened miRNAs targeting IDO1 by using a dual luciferase reporter assay. We tested the function of microRNA-448 (miR-448) by using western blotting (WB) and fluorescence-activated cell sorting (FACS).
Results: We demonstrated that stable IDO1 overexpression enhanced xenograft tumor growth in BALB/c mice but not in BALB/c nude mice. We also revealed the involvement of posttranscriptional regulation of IDO1 in colon cancer by observing IDO1 protein levels and mRNA levels. Furthermore, ectopic expression of miRNA mimics suggested that miR-448 could significantly downregulate IDO1 protein expression. Notably, we proved that miR-448 suppressed the apoptosis of CD8+ T cells by suppressing IDO1 enzyme function.
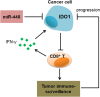
Conclusion: Our findings indicated that IDO1 suppressed the CD8+ T cell response in colon cancer. miR-448, as a tumor-suppressive miRNA, enhanced the CD8+ T cell response by inhibiting IDO1 expression. The results provide a theoretical basis for the development of new immunotherapy for the treatment of colon cancer.
Keywords: CD8; Colon cancer; IDO1; Immunology; Tumor microenvironment; miR-448.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
