The subcellular localization of type I p21-activated kinases is controlled by the disordered variable region and polybasic sequences
- PMID: 31391252
- PMCID: PMC6768646
- DOI: 10.1074/jbc.RA119.007692
The subcellular localization of type I p21-activated kinases is controlled by the disordered variable region and polybasic sequences
Abstract
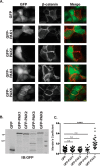
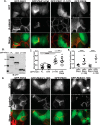
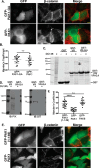
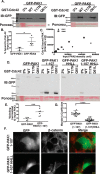
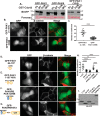
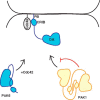
p21-activated kinases (PAKs) are serine/threonine kinase effectors of the small GTPases Rac and Cdc42 and major participants in cell adhesion, motility, and survival. Type II PAKs (PAK4, -5, and -6) are recruited to cell-cell boundaries, where they regulate adhesion dynamics and colony escape. In contrast, the type I PAK, PAK1, does not localize to cell-cell contacts. We have now found that the other type I PAKs (PAK2 and PAK3) also fail to target to cell-cell junctions. PAKs contain extensive similarities in sequence and domain organization; therefore, focusing on PAK1 and PAK6, we used chimeras and truncation mutants to investigate their differences in localization. We observed that a weakly conserved sequence region (the variable region), located between the Cdc42-binding CRIB domain and the kinase domain, inhibits PAK1 targeting to cell-cell junctions. Accordingly, substitution of the PAK1 variable region with that from PAK6 or removal of this region of PAK1 resulted in its localization to cell-cell contacts. We further show that Cdc42 binding is required, but not sufficient, to direct PAKs to cell-cell contacts and that an N-terminal polybasic sequence is necessary for PAK1 recruitment to cell-cell contacts, but only if the variable region-mediated inhibition is released. We propose that all PAKs contain cell-cell boundary-targeting motifs but that the variable region prevents type I PAK accumulation at junctions. This highlights the importance of this poorly conserved, largely disordered region in PAK regulation and raises the possibility that variable region inhibition may be released by cellular signals.
Keywords: Cdc42; adherens junction; cell adhesion; cell signaling; cell-cell contact; intrinsically disordered region; serine/threonine-protein kinase PAK1; serine/threonine-protein kinase PAK6; small GTPase; subcellular localization.
© 2019 Sun et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

