Pol II phosphorylation regulates a switch between transcriptional and splicing condensates
- PMID: 31391587
- PMCID: PMC6706314
- DOI: 10.1038/s41586-019-1464-0
Pol II phosphorylation regulates a switch between transcriptional and splicing condensates
Abstract
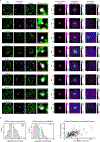
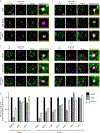
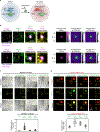
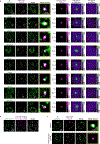
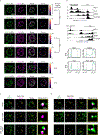
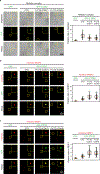
The synthesis of pre-mRNA by RNA polymerase II (Pol II) involves the formation of a transcription initiation complex, and a transition to an elongation complex1-4. The large subunit of Pol II contains an intrinsically disordered C-terminal domain that is phosphorylated by cyclin-dependent kinases during the transition from initiation to elongation, thus influencing the interaction of the C-terminal domain with different components of the initiation or the RNA-splicing apparatus5,6. Recent observations suggest that this model provides only a partial picture of the effects of phosphorylation of the C-terminal domain7-12. Both the transcription-initiation machinery and the splicing machinery can form phase-separated condensates that contain large numbers of component molecules: hundreds of molecules of Pol II and mediator are concentrated in condensates at super-enhancers7,8, and large numbers of splicing factors are concentrated in nuclear speckles, some of which occur at highly active transcription sites9-12. Here we investigate whether the phosphorylation of the Pol II C-terminal domain regulates the incorporation of Pol II into phase-separated condensates that are associated with transcription initiation and splicing. We find that the hypophosphorylated C-terminal domain of Pol II is incorporated into mediator condensates and that phosphorylation by regulatory cyclin-dependent kinases reduces this incorporation. We also find that the hyperphosphorylated C-terminal domain is preferentially incorporated into condensates that are formed by splicing factors. These results suggest that phosphorylation of the Pol II C-terminal domain drives an exchange from condensates that are involved in transcription initiation to those that are involved in RNA processing, and implicates phosphorylation as a mechanism that regulates condensate preference.
Figures






Comment in
-
Switching Condensates: The CTD Code Goes Liquid.Trends Biochem Sci. 2020 Jan;45(1):1-3. doi: 10.1016/j.tibs.2019.10.009. Epub 2019 Nov 13. Trends Biochem Sci. 2020. PMID: 31734037
References
Methods references
-
- tlambert03/LLSpy: v0.3.8. (Zenodo, 2018).
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

