The mRNA degradation factor Xrn1 regulates transcription elongation in parallel to Ccr4
- PMID: 31392315
- PMCID: PMC6765136
- DOI: 10.1093/nar/gkz660
The mRNA degradation factor Xrn1 regulates transcription elongation in parallel to Ccr4
Abstract
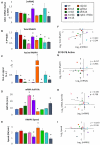
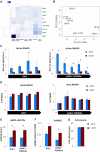
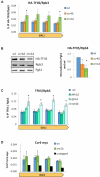
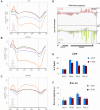
Co-transcriptional imprinting of mRNA by Rpb4 and Rpb7 subunits of RNA polymerase II (RNAPII) and by the Ccr4-Not complex conditions its post-transcriptional fate. In turn, mRNA degradation factors like Xrn1 are able to influence RNAPII-dependent transcription, making a feedback loop that contributes to mRNA homeostasis. In this work, we have used repressible yeast GAL genes to perform accurate measurements of transcription and mRNA degradation in a set of mutants. This genetic analysis uncovered a link from mRNA decay to transcription elongation. We combined this experimental approach with computational multi-agent modelling and tested different possibilities of Xrn1 and Ccr4 action in gene transcription. This double strategy brought us to conclude that both Xrn1-decaysome and Ccr4-Not regulate RNAPII elongation, and that they do it in parallel. We validated this conclusion measuring TFIIS genome-wide recruitment to elongating RNAPII. We found that xrn1Δ and ccr4Δ exhibited very different patterns of TFIIS versus RNAPII occupancy, which confirmed their distinct role in controlling transcription elongation. We also found that the relative influence of Xrn1 and Ccr4 is different in the genes encoding ribosomal proteins as compared to the rest of the genome.
© The Author(s) 2019. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures



References
-
- Timmers H.T.M., Tora L.. Transcript buffering: a balancing act between mRNA synthesis and mRNA degradation. Mol. Cell. 2018; 72:10–17. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

