Epigenetic factors Dnmt1 and Uhrf1 coordinate intestinal development
- PMID: 31394080
- PMCID: PMC6842436
- DOI: 10.1016/j.ydbio.2019.08.002
Epigenetic factors Dnmt1 and Uhrf1 coordinate intestinal development
Abstract
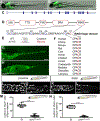
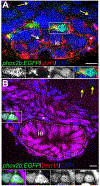
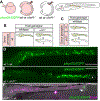
Intestinal tract development is a coordinated process involving signaling among the progenitors and developing cells from all three germ layers. Development of endoderm-derived intestinal epithelium has been shown to depend on epigenetic modifications, but whether that is also the case for intestinal tract cell types from other germ layers remains unclear. We found that functional loss of a DNA methylation machinery component, ubiquitin-like protein containing PHD and RING finger domains 1 (uhrf1), leads to reduced numbers of ectoderm-derived enteric neurons and severe disruption of mesoderm-derived intestinal smooth muscle. Genetic chimeras revealed that Uhrf1 functions both cell-autonomously in enteric neuron precursors and cell-non-autonomously in surrounding intestinal cells, consistent with what is known about signaling interactions between these cell types that promote one another's development. Uhrf1 recruits the DNA methyltransferase Dnmt1 to unmethylated DNA during replication. Dnmt1 is also expressed in enteric neurons and smooth muscle progenitors. dnmt1 mutants have fewer enteric neurons and disrupted intestinal smooth muscle compared to wildtypes. Because dnmt1;uhrf1 double mutants have a similar phenotype to dnmt1 and uhrf1 single mutants, Dnmt1 and Uhrf1 must function together during enteric neuron and intestinal muscle development. This work shows that genes controlling epigenetic modifications are important to coordinate intestinal tract development, provides the first demonstration that these genes influence development of the ENS, and advances uhrf1 and dnmt1 as potential new Hirschsprung disease candidates.
Keywords: DNA methylation; Enteric nervous system; Hirschsprung disease; Intestinal development; Intestinal epithelium; Intestinal smooth muscle.
Copyright © 2019 Elsevier Inc. All rights reserved.
Figures

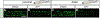






Similar articles
-
Uhrf1 and Dnmt1 are required for development and maintenance of the zebrafish lens.Dev Biol. 2011 Feb 1;350(1):50-63. doi: 10.1016/j.ydbio.2010.11.009. Epub 2010 Nov 30. Dev Biol. 2011. PMID: 21126517 Free PMC article.
-
Nuclear Organization during Hepatogenesis in Zebrafish Requires Uhrf1.Genes (Basel). 2021 Jul 16;12(7):1081. doi: 10.3390/genes12071081. Genes (Basel). 2021. PMID: 34356097 Free PMC article.
-
UHRF1 regulation of Dnmt1 is required for pre-gastrula zebrafish development.Dev Biol. 2016 Apr 1;412(1):99-113. doi: 10.1016/j.ydbio.2016.01.036. Epub 2016 Feb 3. Dev Biol. 2016. PMID: 26851214 Free PMC article.
-
Coordinated Dialogue between UHRF1 and DNMT1 to Ensure Faithful Inheritance of Methylated DNA Patterns.Genes (Basel). 2019 Jan 18;10(1):65. doi: 10.3390/genes10010065. Genes (Basel). 2019. PMID: 30669400 Free PMC article. Review.
-
Thymoquinone Is a Multitarget Single Epidrug That Inhibits the UHRF1 Protein Complex.Genes (Basel). 2021 Apr 22;12(5):622. doi: 10.3390/genes12050622. Genes (Basel). 2021. PMID: 33922029 Free PMC article. Review.
Cited by
-
Regulatory mechanism and biological function of UHRF1-DNMT1-mediated DNA methylation.Funct Integr Genomics. 2022 Dec;22(6):1113-1126. doi: 10.1007/s10142-022-00918-9. Epub 2022 Nov 14. Funct Integr Genomics. 2022. PMID: 36372834 Review.
-
Novel understanding on genetic mechanisms of enteric neuropathies leading to severe gut dysmotility.Eur J Histochem. 2021 Nov 25;65(s1):3289. doi: 10.4081/ejh.2021.3289. Eur J Histochem. 2021. PMID: 34818877 Free PMC article. Review.
-
Who's talking to whom: microbiome-enteric nervous system interactions in early life.Am J Physiol Gastrointest Liver Physiol. 2023 Mar 1;324(3):G196-G206. doi: 10.1152/ajpgi.00166.2022. Epub 2023 Jan 10. Am J Physiol Gastrointest Liver Physiol. 2023. PMID: 36625480 Free PMC article. Review.
-
PGC7 Regulates Genome-Wide DNA Methylation by Regulating ERK-Mediated Subcellular Localization of DNMT1.Int J Mol Sci. 2023 Feb 4;24(4):3093. doi: 10.3390/ijms24043093. Int J Mol Sci. 2023. PMID: 36834503 Free PMC article.
-
Genetic regulation of enteric nervous system development in zebrafish.Biochem Soc Trans. 2024 Feb 28;52(1):177-190. doi: 10.1042/BST20230343. Biochem Soc Trans. 2024. PMID: 38174765 Free PMC article. Review.
References
-
- Amsterdam A, Hopkins N, 2004. Retroviral-mediated insertional mutagenesis in zebrafish. Methods Cell Biol 77, 3–20. - PubMed
-
- Beattie CE, Eisen JS, 1997. Notochord alters the permissiveness of myotome for pathfinding by an identified motoneuron in embryonic zebrafish. Development 124, 713–720. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

