Biomarker Identification Through Integrating fMRI and Epigenetics
- PMID: 31395533
- PMCID: PMC8895412
- DOI: 10.1109/TBME.2019.2932895
Biomarker Identification Through Integrating fMRI and Epigenetics
Abstract
Objective: Integration of multiple datasets is a hot topic in many fields. When studying complex mental disorders, great effort has been dedicated to fusing genetic and brain imaging data. However, an increasing number of studies have pointed out the importance of epigenetic factors in the cause of psychiatric diseases. In this study, we endeavor to fill the gap by combining epigenetics (e.g., DNA methylation) with imaging data (e.g., fMRI) to identify biomarkers for schizophrenia (SZ).
Methods: We propose to combine linear regression with canonical correlation analysis (CCA) in a relaxed yet coupled manner to extract discriminative features for SZ that are co-expressed in the fMRI and DNA methylation data.
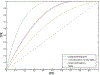
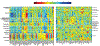
Result: After validation through simulations, we applied our method to real imaging epigenetics data of 184 subjects from the Mental Illness and Neuroscience Discovery Clinical Imaging Consortium. After significance test, we identified 14 brain regions and 44 cytosine-phosphate-guanine(CpG) sites. Average classification accuracy is [Formula: see text]. By linking the CpG sites to genes, we identified pathways Guanosine ribonucleotides de novo biosynthesis and Guanosine nucleotides de novo biosynthesis, and a GO term Perikaryon.
Conclusion: This imaging epigenetics study has identified both brain regions and genes that are associated with neuron development and memory processing. These biomarkers contribute to a good understanding of the mechanism underlying SZ but are overlooked by previous imaging genetics studies.
Significance: Our study sheds light on the understanding and diagnosis of SZ with a imaging epigenetics approach, which is demonstrated to be effective in extracting novel biomarkers associated with SZ.
Figures

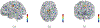
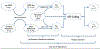
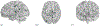
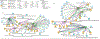

References
-
- Waddington CH, “The epigenotype,” Endeavour, vol. 1, pp. 18–20, 1942.
-
- Abdolmaleky HM et al., “Methylomics in psychiatry: modulation of gene–environment interactions may be through dna methylation,” American Journal of Medical Genetics Part B: Neuropsychiatric Genetics, vol. 127, no. 1, pp. 51–59, 2004. - PubMed
-
- Holliday R. and Pugh JE, “Dna modification mechanisms and gene activity during development,” Science, vol. 187, no. 4173, pp. 226–232, 1975. - PubMed
-
- Riggs AD, “X inactivation, differentiation, and dna methylation,” Cytogenetic and Genome Research, vol. 14, no. 1, pp. 9–25, 1975. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

