Chromatin compartment dynamics in a haploinsufficient model of cardiac laminopathy
- PMID: 31395619
- PMCID: PMC6719452
- DOI: 10.1083/jcb.201902117
Chromatin compartment dynamics in a haploinsufficient model of cardiac laminopathy
Abstract
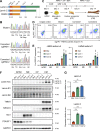
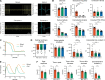
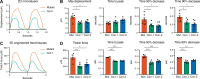
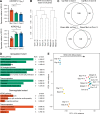
Mutations in A-type nuclear lamins cause dilated cardiomyopathy, which is postulated to result from dysregulated gene expression due to changes in chromatin organization into active and inactive compartments. To test this, we performed genome-wide chromosome conformation analyses in human induced pluripotent stem cell-derived cardiomyocytes (hiPSC-CMs) with a haploinsufficient mutation for lamin A/C. Compared with gene-corrected cells, mutant hiPSC-CMs have marked electrophysiological and contractile alterations, with modest gene expression changes. While large-scale changes in chromosomal topology are evident, differences in chromatin compartmentalization are limited to a few hotspots that escape segregation to the nuclear lamina and inactivation during cardiogenesis. These regions exhibit up-regulation of multiple noncardiac genes including CACNA1A, encoding for neuronal P/Q-type calcium channels. Pharmacological inhibition of the resulting current partially mitigates the electrical alterations. However, chromatin compartment changes do not explain most gene expression alterations in mutant hiPSC-CMs. Thus, global errors in chromosomal compartmentation are not the primary pathogenic mechanism in heart failure due to lamin A/C haploinsufficiency.
© 2019 Bertero et al.
Figures

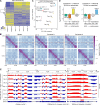
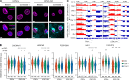

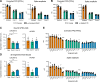
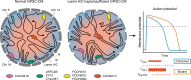
Comment in
-
Challenging the "chromatin hypothesis" of cardiac laminopathies with LMNA mutant iPS cells.J Cell Biol. 2019 Sep 2;218(9):2826-2828. doi: 10.1083/jcb.201907166. Epub 2019 Aug 19. J Cell Biol. 2019. PMID: 31427369 Free PMC article.
References
-
- Adams M.E., Mintz I.M., Reily M.D., Thanabal V., and Bean B.P.. 1993. Structure and properties of omega-agatoxin IVB, a new antagonist of P-type calcium channels. Mol. Pharmacol. 44:681–688. - PubMed
-
- Adriaens C., Serebryannyy L.A., Feric M., Schibler A., Meaburn K.J., Kubben N., Trzaskoma P., Shachar S., Vidak S., Finn E.H., et al. 2018. Blank spots on the map: some current questions on nuclear organization and genome architecture. Histochem. Cell Biol. 150:579–592. 10.1007/s00418-018-1726-1 - DOI - PMC - PubMed
-
- Akinrinade O., Ollila L., Vattulainen S., Tallila J., Gentile M., Salmenpera P., Koillinen H., Kaartinen M., Nieminen M.S., Myllykangas S., et al. 2015. Genetics and genotype-phenotype correlations in Finnish patients with dilated cardiomyopathy. Eur. Heart J. 36:2327–2337. 10.1093/eurheartj/ehv253 - DOI - PMC - PubMed

