The N-terminal region of RTP1S plays important roles in dimer formation and odorant receptor-trafficking
- PMID: 31395660
- PMCID: PMC6779431
- DOI: 10.1074/jbc.RA118.007110
The N-terminal region of RTP1S plays important roles in dimer formation and odorant receptor-trafficking
Abstract
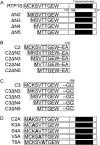
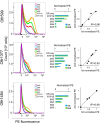
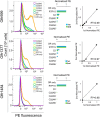
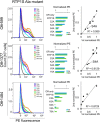
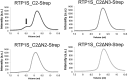
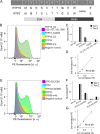
Receptor-transporting protein 1S (RTP1S) is an accessory protein that mediates the transport of mammalian odorant receptors (ORs) into the plasma membrane. Although most ORs fail to localize to the cell surface when expressed alone in nonolfactory cells, functional expression of ORs is achieved with the coexpression of RTP1S. However, the mechanism for RTP1S-mediated OR trafficking remains unclear. In this study, we attempted to reveal the mode of action and critical residues of RTP1S in OR trafficking. Experiments using N-terminal truncation and Ala substitution mutants of RTP1S demonstrated that four N-terminal amino acids have essential roles in OR trafficking. Additionally, using recombinant proteins and split luciferase assays in mammalian cells, we provided evidence for the dimer formation of RTP1S. Furthermore, we determined that the 2nd Cys residue is required for the efficient dimerization of RTP1S. Altogether, these findings provide insights into the mechanism for plasma membrane transport of ORs by RTP1S.
Keywords: G-protein–coupled receptor (GPCR); dimerization; flow cytometry; membrane transport; molecular chaperone; odorant receptors; receptor-transporting protein.
© 2019 Fukutani et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

