Identification of hyper-rewired genomic stress non-oncogene addiction genes across 15 cancer types
- PMID: 31396397
- PMCID: PMC6685999
- DOI: 10.1038/s41540-019-0104-5
Identification of hyper-rewired genomic stress non-oncogene addiction genes across 15 cancer types
Abstract
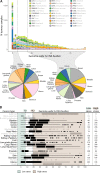
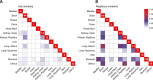
Non-oncogene addiction (NOA) genes are essential for supporting the stress-burdened phenotype of tumours and thus vital for their survival. Although NOA genes are acknowledged to be potential drug targets, there has been no large-scale attempt to identify and characterise them as a group across cancer types. Here we provide the first method for the identification of conditional NOA genes and their rewired neighbours using a systems approach. Using copy number data and expression profiles from The Cancer Genome Atlas (TCGA) we performed comparative analyses between high and low genomic stress tumours for 15 cancer types. We identified 101 condition-specific differential coexpression modules, mapped to a high-confidence human interactome, comprising 133 candidate NOA rewiring hub genes. We observe that most modules lose coexpression in the high-stress state and that activated stress modules and hubs take part in homoeostasis maintenance processes such as chromosome segregation, oxireductase activity, mitotic checkpoint (PLK1 signalling), DNA replication initiation and synaptic signalling. We furthermore show that candidate NOA rewiring hubs are unique for each cancer type, but that their respective rewired neighbour genes largely are shared across cancer types.
Keywords: Cancer; Computational biology and bioinformatics.
Conflict of interest statement
Competing interestsThe authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

