Heterocellular gene signatures reveal luminal-A breast cancer heterogeneity and differential therapeutic responses
- PMID: 31396557
- PMCID: PMC6677833
- DOI: 10.1038/s41523-019-0116-8
Heterocellular gene signatures reveal luminal-A breast cancer heterogeneity and differential therapeutic responses
Abstract
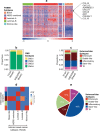
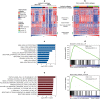
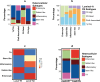
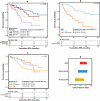
Breast cancer is a highly heterogeneous disease. Although differences between intrinsic breast cancer subtypes have been well studied, heterogeneity within each subtype, especially luminal-A cancers, requires further interrogation to personalize disease management. Here, we applied well-characterized and cancer-associated heterocellular signatures representing stem, mesenchymal, stromal, immune, and epithelial cell types to breast cancer. This analysis stratified the luminal-A breast cancer samples into five subtypes with a majority of them enriched for a subtype (stem-like) that has increased stem and stromal cell gene signatures, representing potential luminal progenitor origin. The enrichment of immune checkpoint genes and other immune cell types in two (including stem-like) of the five heterocellular subtypes of luminal-A tumors suggest their potential response to immunotherapy. These immune-enriched subtypes of luminal-A tumors (containing only estrogen receptor positive samples) showed good or intermediate prognosis along with the two other differentiated subtypes as assessed using recurrence-free and distant metastasis-free patient survival outcomes. On the other hand, a partially differentiated subtype of luminal-A breast cancer with transit-amplifying colon-crypt characteristics showed poor prognosis. Furthermore, published luminal-A subtypes associated with specific somatic copy number alterations and mutations shared similar cellular and mutational characteristics to colorectal cancer subtypes where the heterocellular signatures were derived. These heterocellular subtypes reveal transcriptome and cell-type based heterogeneity of luminal-A and other breast cancer subtypes that may be useful for additional understanding of the cancer type and potential patient stratification and personalized medicine.
Keywords: Cancer genomics; Tumour heterogeneity.
Conflict of interest statement
Competing interestsA.S. has ownership interest as a patent inventor for a patent entitled “Colorectal cancer classification with differential prognosis and personalized therapeutic responses” (patent number PCT/IB2013/060416). A.S. has research funding from Bristol-Myers Squibb and Merck KgaA. M.C.U.C. has a patent: US Patent No. 9,631,239 with royalties paid. The rest of the authors declare that there are no competing interests.
Figures

References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

