Characteristics and Expression Pattern of MYC Genes in Triticum aestivum, Oryza sativa, and Brachypodium distachyon
- PMID: 31398900
- PMCID: PMC6724133
- DOI: 10.3390/plants8080274
Characteristics and Expression Pattern of MYC Genes in Triticum aestivum, Oryza sativa, and Brachypodium distachyon
Abstract
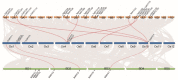
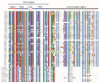
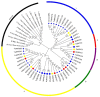
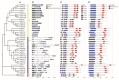
Myelocytomatosis oncogenes (MYC) transcription factors (TFs) belong to basic helix-loop-helix (bHLH) TF family and have a special bHLH_MYC_N domain in the N-terminal region. Presently, there is no detailed and systematic analysis of MYC TFs in wheat, rice, and Brachypodium distachyon. In this study, 26 TaMYC, 7 OsMYC, and 7 BdMYC TFs were identified and their features were characterized. Firstly, they contain a JAZ interaction domain (JID) and a putative transcriptional activation domain (TAD) in the bHLH_MYC_N region and a BhlH region in the C-terminal region. In some cases, the bHLH region is followed by a leucine zipper region; secondly, they display tissue-specific expression patterns: wheat MYC genes are mainly expressed in leaves, rice MYC genes are highly expressed in stems, and B. distachyon MYC genes are mainly expressed in inflorescences. In addition, three types of cis-elements, including plant development/growth-related, hormone-related, and abiotic stresses-related were identified in different MYC gene promoters. In combination with the previous studies, these results indicate that MYC TFs mainly function in growth and development, as well as in response to stresses. This study laid a foundation for the further functional elucidation of MYC genes.
Keywords: Brachypodium distachyon; MYC; bHLH; rice; wheat.
Conflict of interest statement
The authors have declared that they have no conflicts of interest.
Figures


Similar articles
-
Genome-wide analysis of basic helix-loop-helix (bHLH) transcription factors in Brachypodium distachyon.BMC Genomics. 2017 Aug 15;18(1):619. doi: 10.1186/s12864-017-4044-4. BMC Genomics. 2017. PMID: 28810832 Free PMC article.
-
Myc-like transcriptional factors in wheat: structural and functional organization of the subfamily I members.BMC Plant Biol. 2019 Feb 15;19(Suppl 1):50. doi: 10.1186/s12870-019-1639-8. BMC Plant Biol. 2019. PMID: 30813892 Free PMC article.
-
Genomic identification and characterization of MYC family genes in wheat (Triticum aestivum L.).BMC Genomics. 2019 Dec 30;20(1):1032. doi: 10.1186/s12864-019-6373-y. BMC Genomics. 2019. PMID: 31888472 Free PMC article.
-
Global identification, structural analysis and expression characterization of bHLH transcription factors in wheat.BMC Plant Biol. 2017 May 30;17(1):90. doi: 10.1186/s12870-017-1038-y. BMC Plant Biol. 2017. PMID: 28558686 Free PMC article.
-
Basic Helix-Loop-Helix Transcription Factors: Regulators for Plant Growth Development and Abiotic Stress Responses.Int J Mol Sci. 2023 Jan 11;24(2):1419. doi: 10.3390/ijms24021419. Int J Mol Sci. 2023. PMID: 36674933 Free PMC article. Review.
Cited by
-
Metabolic profiling and biomarkers identification in cluster bean under drought stress using GC-MS technique.Metabolomics. 2024 Jul 27;20(4):80. doi: 10.1007/s11306-024-02143-w. Metabolomics. 2024. PMID: 39066988
-
Integrated Analysis of Basic Helix Loop Helix Transcription Factor Family and Targeted Terpenoids Reveals Candidate AarbHLH Genes Involved in Terpenoid Biosynthesis in Artemisia argyi.Front Plant Sci. 2022 Jan 17;12:811166. doi: 10.3389/fpls.2021.811166. eCollection 2021. Front Plant Sci. 2022. PMID: 35111184 Free PMC article.
-
Genome-wide analysis of basic helix-loop-helix genes in Dendrobium catenatum and functional characterization of DcMYC2 in jasmonate-mediated immunity to Sclerotium delphinii.Front Plant Sci. 2022 Aug 2;13:956210. doi: 10.3389/fpls.2022.956210. eCollection 2022. Front Plant Sci. 2022. PMID: 35982703 Free PMC article.
-
Genetic Mapping of Flavonoid Grain Pigments in Durum Wheat.Plants (Basel). 2023 Apr 17;12(8):1674. doi: 10.3390/plants12081674. Plants (Basel). 2023. PMID: 37111897 Free PMC article.
-
Potential role of the regulatory miR1119-MYC2 module in wheat (Triticum aestivum L.) drought tolerance.Front Plant Sci. 2023 May 30;14:1161245. doi: 10.3389/fpls.2023.1161245. eCollection 2023. Front Plant Sci. 2023. PMID: 37324698 Free PMC article.
References
-
- Xu Y.H., Liao Y.C., Lv F.F., Zhang Z., Sun P.W., Gao Z.H., Hu K.P., Sui C., Jin Y., Wei J.H. Transcription Factor AsMYC2 Controls the Jasmonate-responsive Expression of ASS1 Regulating Sesquiterpene Biosynthesis in Aquilaria sinensis (Lour.) Gilg. Plant Cell Physiol. 2017;58:1924–1933. doi: 10.1093/pcp/pcx122. - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

