Generation of recombinant MVA-norovirus: a comparison study of bacterial artificial chromosome- and marker-based systems
- PMID: 31399106
- PMCID: PMC6688233
- DOI: 10.1186/s12985-019-1212-y
Generation of recombinant MVA-norovirus: a comparison study of bacterial artificial chromosome- and marker-based systems
Abstract
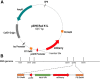
Background: Recombinant Modified Vaccinia Virus Ankara has been employed as a safe and potent viral vector vaccine against infectious diseases and cancer. We generated recMVAs encoding norovirus GII.4 genotype capsid protein by using a marker-based approach and a BAC-based system. In the marker-based approach, the capsid gene together with a reporter gene was introduced into the MVA genome in DF-1 cells. Several rounds of plaque purification were carried out to get rid of the WT-MVA. In the BAC-based approach, recMVA-BAC was produced by en passant recombineering in E. coli. Subsequently, the recMVAs were rescued in DF-1 cells using a helper rabbit fibroma virus. The BAC backbone and the helper virus were eliminated by passaging in DF-1 cells. Biochemical characteristics of the recMVAs were studied.
Results: We found the purification of the rare spontaneous recombinants time-consuming in the marker-based system. In contrast, the BAC-based system rapidly inserted the gene of interest in E. coli by en passant recombineering before virion production in DF-1 cells. The elimination of the reporter gene was found to be faster and more efficient in the BAC-based approach. With Western blotting and electron microscopy, we could prove successful capsid protein expression and proper virus-assembly, respectively. The MVA-BAC produced higher recombinant virus titers and infected DF-1 cells more efficiently.
Conclusions: Comparing both methods, we conclude that, in contrast to the tedious and time-consuming traditional method, the MVA-BAC system allows us to quickly generate high titer recMVAs.
Keywords: Bacterial artificial chromosome; Norovirus; Recombinant MVA; Self-excising.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures




Similar articles
-
Rapid generation of markerless recombinant MVA vaccines by en passant recombineering of a self-excising bacterial artificial chromosome.J Virol Methods. 2010 Sep;168(1-2):233-6. doi: 10.1016/j.jviromet.2010.04.012. Epub 2010 Apr 24. J Virol Methods. 2010. PMID: 20417665
-
Recombination-mediated genetic engineering of a bacterial artificial chromosome clone of modified vaccinia virus Ankara (MVA).PLoS One. 2008 Feb 20;3(2):e1638. doi: 10.1371/journal.pone.0001638. PLoS One. 2008. PMID: 18286194 Free PMC article.
-
Genetic manipulation of poxviruses using bacterial artificial chromosome recombineering.Methods Mol Biol. 2012;890:37-57. doi: 10.1007/978-1-61779-876-4_3. Methods Mol Biol. 2012. PMID: 22688760
-
Construction and isolation of recombinant MVA.Methods Mol Biol. 2004;269:77-100. doi: 10.1385/1-59259-789-0:077. Methods Mol Biol. 2004. PMID: 15114009 Review.
-
[Modified vaccinia virus ankara (MVA)--development as recombinant vaccine and prospects for use in veterinary medicine].Berl Munch Tierarztl Wochenschr. 2015 Nov-Dec;128(11-12):464-72. Berl Munch Tierarztl Wochenschr. 2015. PMID: 26697713 Review. German.
Cited by
-
Comparison of Recombinant MVA Selection Methods Based on F13L, D4R and K1L Genes.Viruses. 2022 Mar 4;14(3):528. doi: 10.3390/v14030528. Viruses. 2022. PMID: 35336935 Free PMC article.
-
Vaccinia Virus: From Crude Smallpox Vaccines to Elaborate Viral Vector Vaccine Design.Biomedicines. 2021 Nov 26;9(12):1780. doi: 10.3390/biomedicines9121780. Biomedicines. 2021. PMID: 34944596 Free PMC article. Review.
-
Viral Vector-Based Chlamydia trachomatis Vaccines Encoding CTH522 Induce Distinct Immune Responses in C57BL/6J and HLA Transgenic Mice.Vaccines (Basel). 2024 Aug 22;12(8):944. doi: 10.3390/vaccines12080944. Vaccines (Basel). 2024. PMID: 39204067 Free PMC article.
-
Expression of an Efficient Selection Marker Out of a Duplicated Site in the ITRs of a Modified Vaccinia Virus Ankara (MVA).Vaccines (Basel). 2024 Dec 6;12(12):1377. doi: 10.3390/vaccines12121377. Vaccines (Basel). 2024. PMID: 39772039 Free PMC article.
-
Sequestration of Late Antigens Within Viral Factories Impairs MVA Vector-Induced Protective Memory CTL Responses.Front Immunol. 2019 Dec 4;10:2850. doi: 10.3389/fimmu.2019.02850. eCollection 2019. Front Immunol. 2019. PMID: 31867011 Free PMC article.
References
-
- Hasing ME, Lee BE, Preiksaitis JK, Tellier R, Honish L, et al. Emergence of a new norovirus GII. 4 variant and changes in the historical biennial pattern of norovirus outbreak activity in Alberta, Canada, from 2008 to 2013. J Clin Microbiol. 2013;51:2204–2211. doi: 10.1128/JCM.00663-13. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

